
Narrative review of bovine milk exosomes as a potential therapeutic option for hepatic fibrosis
Introduction
Hepatic fibrosis (HF) due to viral, toxic or metabolic chronic liver diseases is a major challenge of global health (1). Cirrhosis is currently the 11th most common cause of death in the world and the fourth most frequent cause of death in adults in central Europe (2-4). Progressive accumulation of extracellular matrix (ECM), which destroys the physiological architecture of the liver, is the pathological hallmark of HF (5). Toxic, viral, or metabolic factors lead to hepatocyte damage and infiltration of immune cells that activate trans-differentiation of hepatic stellate cells (HSCs) into collagen-producing myofibroblasts (6-9). HSC activation is considered as a pivotal event in HF (6-10). HSCs are mainly responsible for the excessive accumulation of ECM proteins in the liver associated with increased expression of type I collagen (COL1A1), smooth muscle α-actin (α-SMA) and increased cellular proliferation (10,11).
The liver’s fate to either pass into an anti-fibrotic scar-dissolving stage or to proceed into an uninhibited fibrosis-promoting stage is mainly regulated by non-parenchymal cells, including Kupffer cells and other immune cells (12-16). Hepatocyte apoptosis and release of damage-associated molecular patterns (DAMPs) from hepatocytes activate Toll-like receptor 4 (TLR4), which activates HSCs directly and induces the recruitment and activation of lymphocytes and macrophages that contribute to the promotion of HSC trans-differentiation and myofibroblast activation by producing pro-inflammatory and fibrogenic cytokines (17-19). Distinct macrophage subpopulations on the other hand participate in the resolution of fibrosis through the expression of matrix metalloproteinases (MMPs) (20,21). Thus, on the molecular basis, a complex network of cytokine-induced signaling pathways orchestrates fibrogenic cell interactions. Transforming growth factor-β (TGF-β), platelet-derived growth factor (PDGF), and the inflammasome (NLRP3)-caspase 1 pathway, as well as WNT/β-catenin signaling have been suggested as key signaling pathways associated with HSC activation and fibrosis progression (22-25).
There is recent interest in the role of extracellular vesicles (EVs), in particular exosomes, in the regulation of HSC differentiation and HSC homeostasis. Exosomes are small discoid EVs originating from endosomes that are 30–150 nm in diameter and have an outer lipid bilayer membrane. They participate in immune responses, cell migration, cell differentiation, and tumor invasion and mediate intercellular communication, regulating the biological activity of receptor cells through proteins, nucleic acids including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and lipids as integral constituents (26,27). Exosomes of various origin, i.e., hepatocytes, virally-infected hepatocytes, macrophages, lymphocytes, cholangiocytes, HSCs, adipocyte- and bone marrow-derived mesenchymal stem cells (MSCs), and blood plasma, all interfere with HSC homeostasis, activity and proliferation. Whereas certain exosomes induce HSC-mediated fibrogenesis (28-41), others contribute to the resolution and suppression of fibrogenesis offering new treatment options for HF (42-55). Thus, exosomes are considered as powerful tools for the treatment of HF (55,56), as long as an approved standardized drug for the therapy of HF does not exist (57). Recent evidence underlines the beneficial therapeutic effects of bovine milk-derived exosomes (BMEX) in the treatment of inflammatory bowel diseases and necrotizing enterocolitis (58-63). It is conceivable that BMEX and their miRNA cargo not only improve intestinal but also hepatic functions.
The objective of this narrative review is the presentation of experimental research data for potential anti-fibrotic and fibrotic effects of BMEX and their possible contribution for the alleviation or treatment of HF. We present the following article in accordance with the Narrative Review reporting checklist (available at https://dmr.amegroups.com/article/view/10.21037/dmr-21-79/rc).
Methods
Literature research was performed using the PubMed database between 2000–2021 selecting original research papers in English language including the search items: HF, liver fibrosis, liver cirrhosis, hepatic stellate cell, Ito cell, exosome, fibrosis, fibrogenesis, miRNA (miR), fibrogenic miRNA, anti-fibrogenic microRNA (miRNA, miR), bovine milk exosome (cow milk-derived exosome), bovine milk EV (cow milk-derived EV), milk miRNA, metformin, metformin-induced effects on miRNA expression. Original data of reported biological effects of miRNAs in HSC transformation or HF pathogenesis were extracted and provided in the corresponding miRNA section. Literature data of bovine milk exosome-derived miRNAs were inspected and related to their potential anti-fibrotic and fibrotic effects (Table 1).
Table 1
| Items | Specification |
|---|---|
| Date of search (specified to date, month and year) | December 2021 |
| Databases and other sources searched | PubMed |
| Search terms used (including MeSH and free text search terms and filters) | Exosome; hepatic stellate cell; miRNA, fibrogenesis |
| Timeframe | 2000–2021 |
| Inclusion and exclusion criteria (study type, language restrictions etc.) | Inclusion: Original research articles, English language; Exclusion: articles repeating published data |
| Selection process (who conducted the selection, whether it was conducted independently, how consensus was obtained, etc.) | First author conducted paper selection |
Discussion
Hepatic stellate cells: potential targets of BMEX
BMEX and their miRNAs are bioavailable and reach the systemic circulation as well as peripheral tissues in humans and animal models (64-73). BMEX are regarded as promising biologics to reach distant organs for the treatment of human diseases (cross-species communication) (74-77). When fluorophore-labeled BMEX were administered retro-orbitally, BMEX accumulated in liver, spleen, and lungs in mice (69). Manca et al. (70) confirmed that BMEX labeled with fluorophores or fluorescent fusion proteins accumulated in the liver, spleen and brain following suckling, oral gavage and intravenous administration in mice and pigs. When synthetic, fluorophore-labeled miRNAs were transfected into BMEX and administered to mice, distinct species of miRNAs demonstrated unique distribution profiles and accumulated in the liver and other organs (70). The finding that human vascular endothelial cells transport BMEX by endocytosis is an important step for their delivery to organs and peripheral tissues (69).
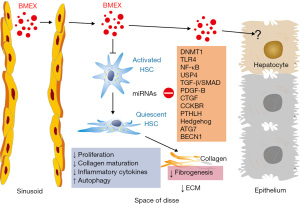
Samuel et al. (73) recently demonstrated that 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindotricarbocyanine iodide (DiR)-labeled bovine milk-derived EVs (30–150 nm, including TSG101-positive EV fractions in the density range of BMEX) preferentially accumulated in the liver of mice 24 h after oral EV gavage. Quantitative proteomic analysis confirmed the presence of bovine-specific proteotypic peptides in liver tissues of mice orally administered with milk-derived EVs confirming their bioavailability in the liver after oral uptake (73). Therefore, it is conceivable that BMEX may enter the space of Disse and approach HSCs (Figure 1). Betker et al. (75) have shown that antibodies are present in exosome preparations, and that DiR-labeled BMEX are absorbed as intact particles from the gastrointestinal tract and accumulated in the liver via the “neonatal” Fc receptor, FcRn. Notably, expression of Fc fragment receptors of IgG has been reported on rat HSCs (78). It has been shown in mice either fed a BMEX-supplemented or BMEX-deficient diet that BMEX supplementation significantly modified hepatic mRNA expression of genes involved in purine metabolism (79). Recently, López de Las Hazas et al. (80) confirmed in a mice model that BMEX-transported miRNAs are stable during gastrointestinal digestion, are bioavailable and reached the liver increasing the expression of miRNA-148a 1 h after BMEX administration. Thus, basic research evidence indicates that BMEX and their transported miRNAs reach the liver and may reach HSCs modifying their posttranslational gene expression.
Milk fat globule-EGF factor 8 in HSC regulation
Recent evidence indicates that milk fat globule-EGF factor 8 (MFG-E8) is a strong inhibitor of activation of human primary HSCs (81). Notably, levels of MFG-E8 are reduced in cirrhotic liver tissue from patients compared with controls (81). MFG-E8, a secretome member of MSCs, is secreted by umbilical cord-derived MSCs (UCMSCs). In mice with fibrosis, the accumulation of ECM proteins was significantly reduced 3 days after injecting secretomes from UCMSCs, and to a greater extent from hepatocyte-like UCMSCs (hpUCMSCs) (81). MFG-E8 down-regulated the expression of TGF-β type 1 receptor by binding to αvβ3 integrin on HSCs. In mice, injection of recombinant human MFG-E8 had anti-fibrotic effects comparable to those of the hpUCMSC secretome, reducing ECM deposition and HSC activation. Co-injection of an antibody against MFG-E8 reduced the anti-fibrotic effects of the hpUCMSC secretome in mice (81). In a mouse model of bleomycin-induced fibrosis and in the TSK mouse model (a genetic model of systemic sclerosis), deficient expression of MFG-E8 significantly enhanced both pulmonary and skin fibrosis, and administration of recombinant MFG-E8 significantly inhibited bleomycin-induced dermal fibrosis (82). In contrast, integrin α(v)-bound MFG-E8 associates with PDGFRB and focal adhesion kinase after PDGF-BB treatment, which results in cell surface retention of PDGFRB, delays receptor degradation and potentiates its downstream signaling (83).
MFG-E8 protein is a major constituent of MFG membranes (84), but is detectable only in minor amounts in milk EVs and BMEX (84-86). In contrast, MFG-E8 mRNA has been detected in BMEX (74) that may thus reach HSCs.
MiRNAs in HSC activation and liver fibrosis
Emerging evidence indicates that miRNAs are abnormally expressed in activated HSCs and play an important role in fibrogenesis (87-89). MiRNAs regulate HSC activation, proliferation, migration and apoptosis, as well as collagen production and ECM deposition (89). MiRNAs are of critical importance in the control of the Hedgehog (Hh) pathway of HSCs (90), which recently reached attention in the pathogenesis of HF (91).
Among various miRNAs studied in association with HSC activation and HF, certain anti-fibrotic miRNAs including miRNA-152, a homolog of miRNA-148a and member of the miRNA-148a/152 family (92), and miRNA-29 family members are down-regulated, whereas the fibrogenic miRNAs 34a/c, miRNA-125b and miRNA-21 were consistently up-regulated in activated HSCs (89).
DNMT1 and miRNA-148a in HSCs and hepatocytes
Epigenetic regulation plays an important role for HSC homeostasis and activation (93-96). Recent evidence indicates that DNA methyltransferase 1 (DNMT1) expression is increased in activated HSCs compared with quiescent HSCs (97). When rat HSC-T6 cells were incubated with PDGF-BB, they exhibited a time-dependent increase in protein expression of DNMT1, α-SMA and COL1A1, respectively, whereas siRNA-treatment of DNMT1 or DNMT1 inhibition with 5-aza-2’-deoxycytidine significantly reduced α-SMA and COL1A1 expression (97). In addition, nuclear p-ERK1/2 expression was significantly down-regulated in DNMT1-siRNA cells and 5′-aza-2′-deoxycytidine treated cultures, which was associated with reduced cell proliferation (97). Moreover, DNMT1 controls the expression of the long noncoding RNA H19 (lncRNA H19) (97), which is dysregulated in HF and is a sponge of miRNA-148a-3p (98). The role of lncRNA H19 in HF is still controversial. One research group observed that lncRNA H19 was significantly down-regulated in HSCs and fibrosis tissues (97,99), whereas other authors reported that cholangiocyte-derived exosomal lncRNA H19 promotes cholestatic liver injury in mice and humans (34), promotes HSC activation and cholestatic HF (36), cholangiocyte proliferation and cholestatic HF in biliary atresia (37). Huang et al. (100) investigated the regulatory effects of the lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) on HF and HSC activation. Up-regulation of NEAT1 and cytohesin 3 (CYTH3) and down-regulation of miRNA-148a-3p and miR-22-3p were observed in mouse fibrotic liver tissues (100). Knockdown of NEAT1 or CYTH3 attenuated HF and collagen deposition in vivo and the activation of HSCs in vitro. MiRNA-148a-3p and miRNA-22-3p inhibitor facilitated HSC activation and collagen fiber expression (100). These data are in accordance with the known posttranslational inhibition of DNMT1 expression via miRNA-148a (101,102). MiRNA-148a-mediated suppression of DNMT1 may thus reduce α-SMA, COL1A1 and nuclear p-ERK1/2 expression (97).
Cytochrome P450 (CYP) enzymes are markedly decreased in alcoholic hepatitis (AH), a critical preclinical inflammatory condition resulting in HF (103). It has been demonstrated by Luo et al. (104) that miRNA-148a promotes CYP2B6 expression by increasing mRNA stability via directly binding to the 3’-UTR sequence of CYP2B6. In contrast, ethanol exposure of hepatocytes via suppressing hepatocyte nuclear factor 4α (HNF4α) decreases the expression of miRNA-148a and thus impairs CYP2B6 expression (104). As shown in cultured hepatocytes and mouse livers, alcohol exposure inhibits forkhead box protein O1 (FoxO1) expression, which correlates with decreased miRNA-148a levels (105). FoxO1 was identified as a further transcription factor for MIR148A transactivation (105). Alcohol-induced decline of miRNA-148a expression in hepatocytes facilitates over-expression of thioredoxin-interacting protein (TXNIP) and NLRP3 inflammasome activation (105). In HepG2 cells, miRNA-148a promotes alcohol dehydrogenase 4 (ADH4) expression by directly binding to the coding sequence of ADH4 and increasing the mRNA stability via an argonaute RISC component 1 (AGO1)-dependent manner (106). Luo et al. (106) suggested that the expressions of key alcohol-metabolizing enzymes are repressed in AH patients, and the non-canonical positive regulation of miRNA-148a on ADH4 reveals a new regulatory mechanism for ADH genes.
It has been shown in neonatal dermal fibroblasts that DNMT1 knockdown suppresses the expression of connective tissue growth factor (CTGF=CCN2) (107), which also plays a key role in the pathogenesis of HF (108,109) and activation of HSCs (110,111).
MiRNAs critically interact with the Hh pathway (90), which plays a pivotal role for HSC activation (91). Patched1 (PTCH1), a negative regulatory factor of the Hh signaling pathway, is down-regulated during HF and associated with hypermethylation of the PTCH1 promoter (112). Notably, salvianolic acid B (Sal B) suppressed the activation of HSCs in CCl4-treated mice and mouse primary HSCs, leading to inhibition of cell proliferation, COL1A1 and α-SMA expression (112). Sal B-mediated up-regulation of PTCH1 was associated with decreased DNA methylation upon Sal B treatment resulting from decreased expression of DNMT1 (112). Interestingly, increased expression of miRNA-152 in Sal B-treated cells was responsible for the hypomethylation of PTCH1 by Sal B. DNMT1 was identified as a direct target of miRNA-152. Further studies showed that a miRNA-152 inhibitor reversed Sal B-mediated PTCH1 up-regulation and DNMT1 down-regulation. DNMT1 is a well-known target of miRNA-148a-3p, miRNA-148b-3p and miRNA-152-3p exhibiting (8mer) highly conserved binding sites (113). These data confirm the importance of the epigenetic DNMT1-mediated regulation of HSC homeostasis.
The uptake of bovine MEX by intestinal epithelial cells (IEC) resulted in an intracellular increase of miRNA-148a associated with a significant suppression of DNMT1 (60,109,110). Notably, miRNA-148a is the most abundant signature miRNA in cow milk and BMEX (114-117) and is highly conserved between mammals (117). The nucleotide stem loop sequence of human (hsa-miR-148a-3p) and bovine (bta-miR-148a-3p) miRNA-148a is identical (118,119). Importantly, miRNA-148a is one of the most highly expressed miRNAs in bovine milk, before and after pasteurization (114,120). These observations imply that the administration of BMEX miRNA-148a may attenuate DNMT1-induced HF.
MiRNA-148a attenuates TGF-β signaling
The TGF-β/SMAD signaling in cooperation with PDGF is considered as one of the most important pathways driving HSC activation and fibrogenesis (24,121). Inactive TGF-β molecules bind to the latency associated protein (LAP) and accumulate in the ECM and must be cleaved by specific proteases to become active. Endothelial cells participate in the conversion of TGF-β from the latent to the active form. Moreover, interactions with transmembrane integrins are considered as the principal activating mechanism for latent TGF-β (122). The active form binds to and activates TGF-β receptor type 2 (TGFBR2), which recruits the TGF-βreceptor type 1 (TGFBR1). The downstream canonical signaling of TGF-β1 converges on SMAD proteins. SMAD3 is crucial for inducing HSC activation and fibrogenic gene transcription such as α-SMA or COL1A1, whereas SMAD2 and SMAD7 are believed to be anti-fibrotic (1,24). However, recent evidence indicates that SMAD2 is up-regulated by endoplasmic reticulum stress related to HSC activation and HF (123). Kupffer cells secrete TGF-β1 promoting HSC activation via the SMAD2/BRD4/C-MYC/EZH2 pathway in HF (124). Furthermore, exosomes derived from human umbilical cord MSCs alleviated HF and decreased the expression of collagen type I and III, TGF-β1 and phosphorylation of SMAD2 (42). Notably, it has been demonstrated in human hepatocellular carcinoma (HCC) cell lines (HepG2, Huh-7, and MHCC97H) that miRNA-148a inhibits the TGF-β/SMAD2 signaling via targeting the SMAD2 3’-UTR decreasing the expression and function of SMAD2 (125).
In accordance with umbilical cord MSC-derived exosomes, BMEX may as well attenuate TGF-β/SMAD2 signaling in HSCs.
MiRNA-148a attenuates cholecystokinin signaling
Dietary fat stimulates growth of pancreatic cancer and promotes fibrosis of the tumor microenvironment through the cholecystokinin (CCK) receptor (CCK-R) (126), whereas a CCK antagonist halts progression of pancreatic cancer precursor lesions and fibrosis in mice (127). Although CCK-Rs have not been reported on HSCs, CCK-R blockade with proglumide could have a similar effect on HSCs as it does on pancreatic stellate cells, thereby preventing or even regressing HF in NASH (128). CCK-Rs have been reported on fibroblasts which when activated, can induce fibrosis (128). CCK-BR expression was markedly up-regulated in the liver and HCC cells of mice fed a saturated fat 75% choline-deficient-ethionine-supplemented (CDE) diet compared with normal hepatic parenchymal cells, whereas CCK-R blockade with proglumide decreased HF associated with NASH (128). In addition, CCK-BR expression was epigenetically suppressed by miRNA-148a (128), which directly targets the mRNA of CCK-BR (129). CCK-BR expression was significantly down-regulated in the Dt81Hepa1-6 cells that over-expressed miRNA-148a, confirming an epigenetic regulatory effect of CCK-BR mRNA expression by miRNA-148a (128). In addition, miRNA-148a was decreased in interstitial renal fibrosis and tubular atrophy (130) underlining the anti-fibrotic effect of miRNA-148a.
Let-7 family miRNAs suppress TGF-β signaling
Lin28 and let-7 play important roles in the regulation in liver diseases (131). The let-7 family of miRNAs is regarded as anti-fibrotic regulators (132). Notably, treatment of HSCs with lipopolysaccharide (LPS) and TGF-β significantly decreased the expressions of let-7a and let-7b. Conversely, over-expression of let-7a and let-7b suppressed the myofibroblastic activation of cultured human HSCs induced by LPS and TGF-β, as evidenced by repressed ACTA2 (α-actin 2), COL1A1, TIMP1 (tissue inhibitor of metalloproteinase 1), and FN1 (fibronectin 1), which supports the view that HSC activation is controlled by let-7 (133). Furthermore, Lin28B deficiency increased the expression of let-7a/let-7b as well as reduced HSC activation and HF in mice with alcoholic liver injury (133). Of note, Lin28 functions as a specific, post-transcriptional inhibitor of let-7 biogenesis (134). High expression of let-7 suppressed epithelial-mesenchymal transition (EMT) and TGF-β signaling (135), which is of key importance in the pathogenesis of HF (136-138). Importantly, let-7 decreases the mRNA expression of TGFBR1, TGF-β receptor type 3 (TGFBR3), and SMAD2 proteins (139,140).
Exosomal let-7 from menstrual blood-derived endometrial stem cells alleviated pulmonary fibrosis (141). Let-7 family members let-7a, let-7b, let-7f, as well as miRNA-148a belong to the most abundant and highly conserved miRNAs of milk EVs of various mammalian species (117). BMEX-derived let-7 and miRNA-148a-mediated suppression of TGF-β signaling may exert anti-fibrogenic activities. In contrast, TGF-β is an outer membrane component of BMEX (142). On the other hand, cow milk-derived EVs contain abundant miRNA-125b (143), which directly targets and inhibits Lin28 mRNA, inhibiting the translation and expression of the Lin28 protein (144), a mechanism that may further enhance let-7-mediated suppression of TGF-β signaling. Recent evidence indicates that the addition of BMEX to IEC18 cells increased the ratio of TGF-β3 to TGF-β1 (145).
MiRNA-148a suppresses TLR4-NFκB signaling
TLR4 enhances TGF-β signaling and plays a crucial role in HF (146-157). Using TLR4-chimeric mice and in vivo LPS challenge, Seki et al. (148) demonstrated that quiescent HSCs, the main precursors for myofibroblasts in the liver, are the predominant target through which TLR4 ligands promote fibrogenesis. In quiescent HSCs, TLR4 activation not only up-regulates chemokine secretion and induces chemotaxis of Kupffer cells, but also down-regulates the TGF-β pseudoreceptor Bambi to sensitize HSCs to TGF-β-induced signals and allow for unrestricted activation by Kupffer cells. LPS-induced Bambi down-regulation and sensitization to TGF-β is mediated by a MyD88-nuclear factor κB (NF-κB)-dependent pathway. Accordingly, Myd88-deficient mice have decreased HF. Thus, modulation of TGF-β signaling by a TLR4-MyD88-NF-κB axis provides a novel link between pro-inflammatory and fibrogenic signals (148). Notably, miRNA-148a inhibits multiple regulatory checkpoints of the TLR4-MyD88-NF-κB axis (62). MiRNA-148a directly targets the 3’-UTR of TLR4 (158). Furthermore, over-expression of miRNA-148a using an agomiR markedly reduced the production of pro-inflammatory cytokines, such as interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) and suppressed NF-κB p65 activation by targeting the TLR4-mediated pathway (158). In addition, miRNA‐148a directly targets calcium/calmodulin‐dependent protein kinase IIα (CaMKIIα) (159), which phosphorylates CARD‐containing MAGUK protein 1 (CARMA1) involved in the activation of IκB kinase α (IKKα)and IκB kinase β (IKKβ). Furthermore, miRNA-148a mitigated hepatic ischemia/reperfusion injury by ameliorating TLR4-mediated inflammation via targeting CaMKIIα in vitro and in vivo (160,161). In addition, miRNA‐148a directly targets IKKα and IKKβ, thereby enhancing the inhibitory effect of IκB on NF‐κB (162). NF-κB directly activates Lin28 transcription and thus rapidly reduces let-7 levels (163).
There is convincing evidence in experimental models of colitis that MEX attenuate TLR4 expression and TLR4-induced NF‐κB activation (62,164,165). Porcine MEX also decreased LPS-induced TLR4/NF-κB signaling pathway activation in IECs (164). Human MEX as well protected against intestinal organoid injury and decreased TLR4 expression (165). It is thus conceivable, that BMEX may also attenuate TLR4-mediated activation of HSCs.
Anti-fibrotic activity of miRNA-29s
PDGF signaling activates HSCs and portal fibroblast proliferation, chemotaxis, migration and cell survival (22,23,166). PDGFB is believed to be the most potent factor associated with early HSCs activation and is transiently increased during the early stage of activation (1,167,168). PDGFR is expressed on the membrane of HSCs and can therefore stimulate HSCs activation through autocrine mechanisms. Rapid induction of PDGFRB is a core feature of HSC activation (167). Depletion of PDGFRB in HSCs decreased injury and fibrosis in vivo, while its auto-activation accelerated fibrosis (167). The miRNA-29 family (miRNA-29a, miRNA-29b-1, miRNA-29b-2, miRNA-29c) is regarded to exert important anti-fibrogenic effects in various tissue fibroses including HF (169-176). In the healthy liver, miRNA-29 is highly expressed in hepatic HSCs and is responsible for repression of ECM proteins, in particular collagens, as well as PDGF-C and insulin-like growth factor 1 (IGF-1) (169). During fibrogenesis, fibrogenic growth factors are increased and stimulate HSCs in a paracrine and autocrine manner. Fibrogenic stimulation of TGF-β and PDGF-BB results in miRNA-29 repression. The resulting loss of miRNA-29 by TGF-β and PDGF-BB results in the abolished repression of fibrogenic expression of ECM, PDGF-C or IGF-1. Enhanced secretion of PDGF-C and IGF-1 stimulates HSC in an autocrine manner increasing HSC proliferation and ECM production (169). IGF-1 is a strong mitogen stimulating phosphatidylinositol 3-kinase (PI3K)/AKT signaling. Thus, the loss of miRNA-29 is regarded to be due to the response of HSCs to the exposure to the fibrogenic mediators TGF-β and PDGF-BB (169). Kwiecinski et al. (175) provided evidence that myofibroblastic transition of primary HSCs resulted in the loss of miRNA-29, but in a significant increase of PDGF-C and IGF-1. Compensation of reduced miRNA-29 levels by miRNA-29 over-expression in myofibroblastic HSCs was followed by a definitive repression of IGF-1 and PDGF-C synthesis (175). In contrast, Sanz et al. (176) reported that targeted over-expression of IGF-1 by activated HSCs restricts their activation, attenuates fibrogenesis, and accelerates liver regeneration.
Notably, BMEX enhanced IEC expression of glucose-regulated protein 94 (GRP94) (177). GRP94 is a critical endoplasmic reticulum (ER) chaperone that enhances the secretion of properly folded mature IGF-1 (178).
Fu et al. (179) recently showed that miRNA-29a-3p suppresses HF pathogenesis by modulating HSC proliferation via targeting PIK3R3 gene (phosphatidylinositol 3-kinase, regulatory subunit 3) modulating PI3K/AKT signaling. In miRNA-29a transgenic mice, miRNA-29a alleviated bile duct ligation with exacerbation of HF in mice through suppression of DNMT1 and DNMT3B lowering COL1A1 and SET domain containing 1A (SET1A) expression in HSCs (180). Gain of miRNA-29a signaling resulted in DNA hypomethylation and high PTEN expression. in vitro, miRNA-29a mimic transfection lowered COL1A1, DNMT1, DNMT3B and SET1A expression in HSCs (180). Thus, epigenetic action of miRNA-29a ameliorates the development of cholestatic HF. There is further evidence that miRNA-29a disrupts DNMT3B to ameliorate diet-induced non-alcoholic steatohepatitis (NASH) in mice (181). In accordance, a miRNA-29a mimic transfection reduced DNMT3B expression in primary HSCs (181).
An up-regulation of miRNA-29 and suppression of HSC activation has also been observed by class II histone deacetylase (HDAC) inhibition (182). In miRNA-29a transgenic mice, over-expression of miRNA-29a decreased COL1A1, histone deacetylases 4 (HDAC4) and activated HSC markers of glial fibrillary acidic protein expression compared to wild-type littermates. Over-expression of miRNA-29a and HDAC4 RNA-interference decreased the expression of fibrotic genes, HDAC4 signaling, and HSC migration and proliferation. In contrast, knockdown of miRNA-29a with an antisense inhibitor increased HDAC4 function, restored HSC migration, and accelerated HSC proliferation (183). Hepatic tissue in miRNA-29a transgenic mice displayed a weak fibrotic matrix concomitant with low fibrotic COL1A1 expression within the affected tissues compared to the wild-type mice fed a high-fat diet (184).
In comparison with patients without HF, miRNA-29b-3p level was remarkably reduced in those with HF (185). TGF-β1-induced HSC activation significantly decreased miRNA-29b-3p expression. However, miRNA-29b-3p over-expression repressed TGF-β1-induced COL1A1 protein and α-SMA expression. Furthermore, it was noted that miRNA-29b-3p directly binds to signal transducer and activator of transcription 3 (STAT3) and suppressed its expression (185). Wang et al. (186) demonstrated that miRNA-29b was significantly down-regulated in human and mice fibrotic liver tissues and in primary activated HSCs. MiRNA-29b down-regulation was directly mediated by SMAD3 through binding to the promoter of MIR29B in the HSC line LX1, whilst miRNA-29b could in turn suppress SMAD3 expression (186). Furthermore, miRNA-29b prevents liver fibrogenesis by inhibiting HSC activation and inducing HSC apoptosis through inhibiting PI3K/AKT signaling. These results provide further insights for the anti-fibrotic effect of miRNA-29b (186). Both miRNA-29b over-expression and SMAD3 silencing antagonized the effects of TGF-β1 on the expression of α-SMA and COL1A1 (187). Furthermore, infection with miRNA-29b mimics suppressed SMAD3 and TGF-β1 expression, suggesting that miRNA-29b inhibited LX-2 activation mediated by both SMAD3 and TGF-β1. Thus, Liang et al. (187) suggested a crosstalk between miRNA-29b and TGF-β1/SMAD3 during LX-2 activation. Homeobox transcript antisense RNA (HOTAIR), is an intergenic long non-coding RNA (lncRNA), which is up-regulated in HSCs in HF and is a target of miRNA-29b. HOTAIR epigenetically modulates PTEN expression via miRNA-29b, a further mechanism promoting HF (188). Transfection of a miRNA-29b precursor markedly attenuated the expression of COL1A1 and COL1A2 mRNAs and additionally blunted the increased expression of α-SMA, DDR2, FN1, ITGB1, and PDGFRB, which are key genes involved in the activation of HSCs (189). Over-expression of miRNA-29b led HSCs to remain in a quiescent state (189). Remarkably, the nucleotide sequences of human PDGFB 3'-UTR (UGGUGCU) and bovine PDGFB 3'-UTR (UGGUGCU) as well as the seed sequences of human miRNA-29b-3p (ACCACGA) and bovine miRNA-29b-3p (ACCACGA) are identical (190,191). Human and bovine let-7-5p are further miRNAs (7mer-m8) with higher probability for targeting human and bovine PDGFB (190,191). Furthermore, PDGFRB is a reported target of miRNA-29-3p (192). Thus, miRNA-29b effectively targets PDGFB-PDGFRB signaling, which represents a key mechanism activating HSCs. To make use of the anti-fibrotic activity of miRNA-29b as a novel miRNA tool, a MRI-visible nanocarrier system targeting HSCs has been developed (193). This pH-sensitive and vitamin A (VA)-conjugated copolymer VA-polyethyleneglycol-polyethyleneimine-poly(N-(N’,N’-diisopropylamino-ethyl)-co-benzyl-amino) aspartamide (T-PBP) micelle efficiently transports the miRNA-29b and miRNA-122 to HSC in a magnetic resonance imaging-visible manner, resulting in a synergistic anti-fibrosis effect via down-regulating the expression of fibrosis-related genes, including COL1A1, α-SMA, and TIMP1 (193).
CTGF (also known as CCN2) is critically involved in HSC activation and HF (108,109,111,194). Notably, fibrogenic signaling is suppressed in HSCs through targeting of CTGF by cellular or exosomal miRNA-199a-5p (195). The basic helix-loop-helix transcription factor, Twist1, binds to the promoter of MIR214 and drives miRNA-214 expression resulting in the suppression of CTGF (29). MiRNA-214 in LX-2 cells was shuttled by exosomes to recipient LX-2 cells or human HepG2 hepatocytes, resulting in suppression of CTGF and its downstream targets, α-SMA and collagen (196). Exosomal transfer of miRNA-214 thus regulates CTGF-dependent fibrogenesis and identifies fibrotic pathways as targets of intercellular regulation by exosomal miRNAs (196). MiRNA-214-enriched exosomes derived from endometrial stromal cells inhibited endometriosis fibrosis (197). Of note, miRNA-29b treatment exhibited an inhibitory effect against skin scar formation via inhibition of the TGF-β1/SMAD/CTGF signaling pathway (198). In accordance, miRNA-29b inhibited endometrial fibrosis by regulating the Sp1-TGF-β1/SMAD-CTGF axis in a rat model (199). Thus, miRNA-29-mediated suppression of CTGF may also attenuate HF. Remarkably, CTGF binds to fibroblast growth factor receptor 2 (FGFR2) and modulates its signaling (200). FGFR2-IIIb is exclusively expressed by hepatocytes, but not by activated HSCs (201).
Runt-related transcription factor 1 (RUNX1) and RUNX2 are fibrogenic transcription factors that are induced at the post-transcriptional level during activation of HSC and suppress TIMP1 via interaction with UTE-1, a regulatory DNA motif on the TIMP1 promoter (202). In the CCl4-induced rat cirrhotic model, selective p38 mitogen-activated protein kinase (MAPK) inhibition exerted salutary effect on liver cirrhosis through down-regulation of RUNX2 (203). However, Baier et al. (64) demonstrated that the expression of RUNX2 increased by 31% in blood mononuclear cells 6 h after consumption of commercial cow milk in adult healthy volunteers compared with baseline. During osteoblast differentiation, miRNA-29b decreased the activity of COL1A1, COL5A3, and COL4A2 3’-UTR sequences in reporter assays, as well as endogenous gene expression but increased RUNX2 expression (204).
In contrast, miRNA-29b targets and negatively regulates parathyroid hormone-like hormone (PTHLH) (205), which is involved in the pathogenesis of HF (206,207). In cultured human HSCs, mRNA and protein levels of α-SMA, COL1A1, MMP-2, and TGF-β1 were increased by PTHLH treatment. A similar increasing pattern was also observed in LX-2 cells. Moreover, PTHLH significantly increased TGF-β1 secretion in cultured media from HSCs (207). Recent evidence links PTHLH to Hh pathway activation (208). PTHLH-mediated parathyroid hormone 1 receptor (PTH1R)-protein kinase C θ (PKC θ) pathway activation was identified as key event in the pro-fibrotic Hh-dependent activation of HSCs (208). As shown in IECs, PTHLH-PTH1R signaling can propagate protein kinase A (PKA) pathway activating downstream nuclear transcription factors, including RUNX2 (209). Thus, miRNA-29b-mediated suppression of PTHLH may suppress RUNX2 expression in HSCs.
Recently, autophagy has attracted much attention in HF pathogenesis. However, the role of autophagy in the course of HF is still controversial (210). Long non-coding small nucleolar RNA host gene 7 (lncRNA-SNHG7) was significantly increased in liver tissue and HSCs of liver fibrosis model of mice, whereas inhibition of SNHG7 expression in liver fibrosis mice could reduce HF (211). SNHG7 binds to miRNA-29b in HSCs and inhibits its expression (211). SNHG7 controls the expression of DNMT3A, which is a downstream target of miRNA-29b. In TGF-β-stimulated normal HSCs, knockdown of SNHG7 expression could restrain DNMT3A and HSCs activation factors α-SMA, COL1α and autophagy-related factors LC3I/II and Beclin1 (211).
Cow milk contains bovine miRNA-29b (64,212). A dose-dependent increase in miRNA-29b plasma levels has been observed with a peak maximum at 6 h after intake of commercial milk by healthy human volunteers (64). MiRNA-29 family members (miRNA-29a, miRNA-29b, miRNA-29c and miRNA-29d) have all been detected in BMEX and milk-derived EVs (73,213,214). It is thus conceivable that BMEX miRNA-29s may reach HSC after oral administration and exert anti-fibrogenic activities.
MiRNA-223
Hepatocyte TAZ/WWTR1 promotes inflammation and fibrosis in NASH (215). A key mechanism linking hepatocyte TAZ to NASH fibrosis is TAZ/TEA domain (TEAD)-mediated induction of Indian hedgehog (Ihh), a secretory factor that activates fibrogenic genes in HSCs (215). It has recently been shown that exosomes enriched in miRNA-223 attenuate non-alcoholic fatty liver disease (NAFLD)-associated fibrosis (54). Furthermore, miRNA-223, which is highly expressed in exosomes of natural killer (NK) cells inhibited TGF-β-induced HSC activation (210). Autophagy-related 7 (ATG7) was confirmed as a direct target of miRNA-223. ATG7 over-expression in LX-2 cells abolished the suppressive effect of NK-derived exosomes on HSC activation (216). Inhibition of autophagy via miRNA-96-5p targeting ATG7 has also been shown to prevent HSC activation (217). It has been shown that autophagy releases lipid species that promote fibrogenesis by activated HSCs in mice and in human tissues (218). In addition, miRNA-223 targets the transcriptional activator with PDZ-binding motif (TAZ), a well-known factor promoting NASH fibrosis (54). After activation, neutrophils can deliver miRNA-223 via exosomes into macrophages (219,220) or hepatocytes (221) thereby inhibiting NASH and HF. Hepatic expression of miRNA-223 targets fibrotic genes including TAZ/WWTR1, NLRP3, IGF1R, and CXCL10 (54,221,222). Transcription factor TAZ/WWTR1 is increased in mouse and human NASH and silencing TAZ can prevent or reverse NASH features, notably fibrosis (218). Interestingly, Liu et al. (223) reported that up-regulation of circular RNA PWWP2A promotes HF via sponging miRNA-223 and miRNA-203. Circ-PWWP2A was up-regulated in both TGF-β- and LPS-activated HSCs and in mouse fibrotic liver tissues and was positively correlated with HSC activation and proliferation (223). In addition, FGFR2 is a direct target of miRNA-223 (224,225).
Izumi et al. (226) reported that miRNA-223 belongs to the group of dominant immune development-related miRNAs of cow milk. Benmoussa et al. (143) found miRNA-223 in BMEX but more abundant quantities in 200 nm EVs sedimenting at a centrifugation speed lower than that for BMEX. They observed that incubation of bovine milk EVs with human cultured HeLa cells led to cellular enrichment in miRNA-223, which was concomitant with decreased expression of a reporter gene placed under the control of miRNA-223, thereby demonstrating the functionality of miRNA-223. Their results suggest that bovine milk EVs transfer their miRNAs to human cells and regulate recipient cell gene expression (227).
Thus, miRNA-223 can be added to the list of potential anti-fibrogenic miRNAs of bovine milk EVs.
MiRNA-125b
You et al. (228) observed increased expression of miRNA-125b in activated HSCs but not in hepatocytes. Inhibition of miRNA-125b suppressed the expression of fibrogenic genes in culture-activated primary HSCs and reduced the basal TGF-β-induced α-SMA expression and cell contraction of the immortalized HSC cell line, whereas ectopic expression of miRNA-125b promoted α-SMA expression and HSC contraction (228). Antagonizing miRNA-125b in vivo significantly alleviated HF in CCl4-treated mice. Mechanistically, over-expression of miRNA-125b in HSCs enhanced RhoA activity by directly targeting StAR-related lipid transfer (START) domain containing 13 (STARD13), a RhoA-specific GTPase-activating protein, whereas knockdown of miRNA-125b abrogated RhoA activation (228). Furthermore, inhibition of RhoA or its downstream molecules, MRTF-A and SRF, attenuated the miRNA-125b-induced α-SMA expression and HSC contraction. Thus, these findings identify a miRNA-125b-STARD13-RhoA-α-SMA signaling cascade in HSCs promoting HF.
In contrast, Hyun et al. (229) reported that miRNA-125b attenuated Hh signaling and promoted liver regeneration by chorionic plate-derived MSC (CP-MSC)-derived exosomes loaded with miRNA-125b. Notably, miRNA-125b is highly expressed in CP-MSCs, but not in LX2 HSCs. MiRNA-125b targeting SMO (Smoothened, frizzled class receptor) was retained in exosomes of CP-MSCs, whereas CP-MSCs with miRNA-125b inhibitor failed to attenuate the expression of Hh signaling and fibrotic genes in the activated HSCs. Therefore, the authors concluded that exosomal miRNA-125b derived from CP-MSCs suppressed the activation of Hh signaling and contributed to liver regeneration. Zhou et al. (230) reported that miRNA-125b directly targets SMAD2 and SMAD4, which inhibits EMT process in HCC cells. Because EMT is closely associated with HSC activation, it is possible that miRNA-125b exerts its anti-fibrotic effect through targeting SMAD4 and other EMT-related genes (230), including Hh signaling, in CCl4-induced HF (90). FGFR2 was identified as a direct target suppressed by miRNA-125b (231,232).
Bovine milk-derived EVs and BMEX, which contain miRNA-125b (143), thus exert controversial effects in the pathogenesis of HF.
MiRNA-155
Zhu et al. (89) reported that miRNA-155 is up-regulated in HF, whereas Kitano et al. (88) provided evidence for reduced expression of miRNA-155 in activated HSCs. According to Csak et al. (233), miRNA-155 affects the progression of HF by regulating the SMAD3 signaling pathway. In particular, miRNA-155 deficiency attenuates steatosis and fibrosis in a mouse model of steatohepatitis (233). In a rat model of AH, miRNA-155 via targeting suppressor of cytokine signaling 1 (SOCS1) activated MAPK signaling and promoted HSC viability and cycle progression and reduced cell apoptosis by silencing SOCS1, whereas inhibition of miRNA-155 up-regulated SOCS1 and inactivated the MAPK signaling pathway, thereby inhibited the proliferation of alcoholic HSCs (234). Bala et al. (235) observed that fibrogenic genes were attenuated in miRNA-155 KO mice after alcohol diet or CCl4 treatment. Furthermore, they showed that TLR4 signaling regulates miRNA-155 as TLR4 KO mice showed no induction of miRNA-155 after alcohol exposure (235). Dai et al. (236) observed that miRNA-155 attenuates activation of HSCs by simultaneously preventing EMT and ERK1 signaling. Bone morphogenetic protein 9 (BMP9) is the most recently discovered member of the BMP family involved in HF (237). Both, RUNX2 and bone morphogenetic protein receptor type 2 (BMPR2) are two direct target genes of miRNA-155 (238). Accordingly, over-expressed miRNA-155 in MSCs reduced the expression of RUNX2 (238).
Both bovine colostrum and mature cow milk contain immune-modulating miRNA-155 (226). Pre-treatment of IEC-6 cells with BMEX increased intracellular miRNA-155 levels (239). Notably, miRNA-155 was enriched in exosomes released from HCC cells, suppressed the expression of PTEN and stimulated HCC cell proliferation (240). In fact, miRNA-155-5p promoted proliferation, invasion and migration, but inhibited apoptosis in HCC by directly targeting the 3’-UTR of PTEN (241). Furthermore, hepatitis C virus (HCV)-induced up-regulation of miRNA-155 promotes hepatocarcinogenesis by activating WNT signaling (242). In addition, miRNA-155 mediates hepatitis B virus (HBV) replication by reinforcing SOCS1 signaling-induced autophagy (243).
These data indicate, that BMEX-derived miRNA-155 may exert unfavorable effects especially in states of HF associated with HBV- and HCV infection and HCC.
MiRNA-30a
TGF-β1 down-regulated the expression of miRNA-30a in activated HSCs and LX-2-exosomes, whereas over-expression of miRNA-30a suppressed α-SMA, TIMP-1, COL1 expression, and cell viability in HSCs (244). MiRNA-30a was significantly down-regulated in HF mice and over-expression of miRNA-30a prevented BDL-induced fibrogenesis, concomitant with the down-regulation of ECM. MiRNA-30a inhibited HSCs autophagy via targeting its downstream effector beclin1 (244). Furthermore, miRNA-30a directly targets the mRNA of growth/differentiation factor 2 (GDF2=BMP9) (245). It has been shown in murine multilineage cells that BMP9, a fibrinogenic transcription factor (237), enhanced the expression of miRNA-21 (246).
MiRNA-30a belongs to the top 50 miRNAs expressed in whey-derived BMEX (213) and counterbalance fibrogenic miRNA-21 signaling.
MiRNA-34a
MiRNA-34a as well as miRNA-155 have been associated with the pathogenesis of NAFLD (247). Li et al. (248) confirmed a direct interaction of peroxisome proliferator-activated receptor-γ (PPARγ) with miRNA-34a and miRNA-34c. Notably, the expression of PPARγ was negatively correlated with the expression of miRNA-34a and miRNA-34c during the activation of HSCs. In activated human HSCs, inhibitors of miRNA-34a and miRNA-34c up-regulated the expression of PPARγ and down-regulated the expression of α-SMA (248). These data suggested that the miRNA-34 family may be involved the process of HF by targeting PPARγ. Furthermore, miRNA-34a appears to play an important role in the process of HF by targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) ACSL1 (249). MiRNA-34a-silenced HSCs showed higher ACSL1 and lower α-SMA, COL1A1, and desmin expression than that of matched negative controls and non-transfected cells (249). Tian et al. (250), using a CCl4-induced rat HF model found that the miRNA-34a/SIRT1/p53 signaling pathway was activated in hepatocytes but not in HSCs. They suggested that the activation of this pathway in hepatocytes results in the apoptosis of hepatocytes and thus activates HSCs (250).
In contrast, Feili et al. (251) observed decreased expression of miRNA-34a-5p in patients with HBV-activated HF and HCC, as well as in CCl4-induced HF model mice. The TGF-β1/SMAD3 pathway was significantly augmented in CC14-induced mice compared with normal control, whereas an inhibitor of TGF-β1 significantly attenuated HF and TGF-β1/SMAD3 activation. Administration of the miRNA-34a-5p mimic de-activated TGF-β1/SMAD3 pathway in human HSCs (LX-2). Moreover, the target gene for miRNA-34a-5p, SMAD4, was predicted and verified in LX-2 cells. Taken together, these data demonstrated that over-expression of miRNA-34 in HSCs ameliorated the development and progression of HF by targeting SMAD4 and regulating TGF-β1/SMAD3 pathway (251). Liu et al. (252) reported that miRNA-34a and miRNA-29c were significantly reduced and Sirt1 protein was increased in PU box binding protein PU.1+/- HSCs compared with WT HSCs. Notably, PU.1 directly binds to both the promoter regions of miRNA-34a and miRNA-29c. PU.1 suppresses Sirt1 translation via transcriptional promotion of miRNA-34a and miRNA-29c, thus promoting Sirt1-mediated HSC activation and thioacetamide-induced HF.
In comparison to bovine colostrum, mature milk contains lower quantities of mRNA-34a (226). Gao et al. (253) presented evidence that under hypoxic conditions BMEX decreased hypoxia inducible factor-1α (HIF-1α). However, bovine miRNA-34a was found to be an effective regulator for alleviating hypoxic injury of IEC-6 via up-regulation of HIF-1α (254). HIF-1α was significantly increased in hepatic fibrotic tissues and activated HSCs. Furthermore, knockdown of HIF-1α expression inhibited the proliferation and activation of HSCs. In addition, HIF-1α-dependent genes and the extensive network of signaling cascades focus on HIF-1α have been reported to associate with the development of HF, suggesting that HIF-1α might play a crucial role in HF (255,256).
BMEX-mediated suppression of HIF-1α may thus exert beneficial effects in HF.
Fibrogenic miRNA-21
MiRNA-21 is up-regulated in fibrogenesis. Wei et al. (257) used PDGF-BB to stimulate HSCs (LX-2 cells) and found that the levels of COL1A1 and α-SMA were reduced while miRNA-21 levels were increased. Remarkably, decreasing the expression of miRNA-21 by transfecting a miRNA-21 inhibitor into LX-2 cells prevented PDGF-BB induced LX-2 cell activation (257). The mRNA level of PTEN was reduced in LX-2 cells by over-expression of miRNA-21, leading to activation of AKT signaling (257). The authors concluded that miRNA-21 induced HSC activation by regulating the PTEN/AKT signaling pathway (257). Moreover, the auto-regulatory feedback loop of miRNA-21/programmed cell death protein 4 (PDCD4)/activation protein-1 (AP-1) was considered a driving force for the development of HF (257). Disrupting this feedback loop by using either miRNA-21- or AP-1 inhibitors significantly diminished fibrogenesis while blocking PDCD4 enhanced fibrogenesis in HSCs (258). MiRNA-21 also modulates extracellular signal-regulated kinase 1 (ERK1) during fibrogenesis by regulating HNF4α and Sprouty 2 (SPRY2) expression (259). Methyl helicterate (MH) has been reported to have protective effects against CCl4-induced hepatic injury and fibrosis in rats. Huang et al. (260) reported that MH treatment significantly decreased miRNA-21 expression and inhibited the activation of the ERK and TGF-β1/SMAD2/3 pathways in liver tissues. MH strongly inhibited HSC-T6 cell activation and reduced collagen accumulation. Notably, miRNA-21 over-expression significantly promoted HSC-T6 cell proliferation, reduced HSC apoptosis, and increased collagen formation, while these abnormal changes induced by miRNA-21 over-expression were significantly reversed by MH treatment. Over-expressed miRNA-21 activated the ERK and TGF-β1/SMADs pathways via repressing SPRY2 and SMAD7, respectively (261). In addition, 3,3’-diindolylmethane (DIM), a natural autolytic product in plants that can down-regulate miRNA-21 expression, suppressed the central TGF-β signaling pathway underlying HSC activation by down-regulating miRNA-21. Decreased miRNA-21 expression was achieved by inhibiting the activity of the transcription factor AP-1 (261). Of note, miRNA-21 is transcriptionally up-regulated in response to SMAD3 rather than SMAD2 activation after TGF-β stimulation. In addition, TGF-β promotes miRNA-21 expression by formation of a microprocessor complex containing SMAD proteins. Elevated miRNA-21 may then act as a fibrogenic miRNA by its repression of the TGF-β inhibitory SMAD7 protein (169). Anti-miRNA-21 treatment reduced liver tumor growth and prevented tumor development. These effects were accompanied with a decrease in HF and a concomitant reduction of CD24+ liver progenitor cells and S100A4+ cancer-associated stromal cells (262). In contrast, Caviglia et al. (263) recently reported that miRNA-21 antisense inhibition did not suppress the activation of murine or human HSCs in culture or in liver slices. Moreover, genetic deletion of miRNA-21 in two independently generated knockout mice or miRNA-21 antisense inhibition did not alter HSC activation or HF in models of toxic and biliary liver injury. As inhibition of the most up-regulated miRNA did not affect HSC activation, HF, or fibrosis-associated liver cancer. Remarkably, Dicer deletion, which decreased HSC miRNA expression only exerted negligible effects on HSC activation and HF (263). On the other hand, Ning et al. (264) reported that miRNA-21 mediates angiotensin II-activated NLRP3 inflammasome and resultant HSC activation via targeting SPRY1 and SMAD7. Hypoxia-responsive miRNA-21-5p promoted RUNX2 expression (at least in part) by targeting the 3’-UTR and down-regulating SMAD7 expression (265), which represses RUNX2 expression (266).
Makhmudi et al. (267) showed that miRNA-21 expression was significantly increased in patients with biliary atresia compared to controls, whereas miRNA-21 expression was significantly lower in cirrhosis than non-cirrhosis patients with biliary atresia. Up-regulated expression of miRNA-21 in liver samples from biliary atresia patients negatively correlated with PTEN expression and downstream activated AKT pathway provoked HF by enhancing α-SMA levels (268).
HCC cells exhibit a great capacity to convert normal HSCs to cancer-associated fibroblasts (CAFs). They secrete exosomal miRNA-21 that directly targeted PTEN, leading to activation of PDK1/AKT signaling in HSCs (269). Knockout of miRNA-21 attenuates AH through the VHL/NF-κB signaling pathway in HSCs (270). Recent evidence indicates that miRNA-21, acting via the HIF-1α/VEGF signaling pathway, is also involved in arsenite-induced HF through mediating aberrant cross-talk of hepatocytes and HSCs (271). In addition, oncogenic virus (HBV, HCV, HPV, and EBV) infections up-regulated host miRNA-21 to evade host immune responses and to promote viral replication (272). Taken together, accumulated evidence supports adverse effects of miRNA-21 in HSC activation, HF and development of HCC.
MiRNA-21 is a dominant miRNA (signature RNA) of commercial cow milk (116) and component of BMEX (58,72,273). In human volunteers, increased plasma miRNA-21 levels have been reported 6 h after milk consumption (68). Table 2 summarizes the anti-fibrogenic and fibrogenic miRNAs in relation to prominent signature miRNAs transported by BMEX.
Table 2
| miRNAs | Effect on HSCs | Mode of action | References |
|---|---|---|---|
| miRNA-148a | Anti-fibrogenic | Inhibition of DNMT1 | Yang et al. (97) |
| miRNA-148a | Anti-fibrogenic | Inhibition of SMAD2 | Jiang et al. (125) |
| miRNA-148a | Anti-fibrogenic | Inhibition of CCKBR | Tucker et al. (128) |
| miRNA-148a | Anti-fibrogenic | Inhibition of TLR4 | Jiang et al. (158) |
| miRNA-148a | Anti-fibrogenic | Inhibition of CAMK2A | Liu et al. (159) |
| miRNA-148a | Anti-fibrogenic | Inhibition of IKBKB | Patel et al. (162) |
| let-7 | Anti-fibrogenic | Inhibition of TGFBR1 and TGFRB3 | Bronevetsky et al. (139) |
| let-7 | Anti-fibrogenic | Inhibition of SMAD2 | Shen et al. (140) |
| miRNA-29a | Anti-fibrogenic | Inhibition of DNMT1 | Yang et al. (180) |
| miRNA-29a | Anti-fibrogenic | Inhibition of DNMT3B | Yang et al. (181) |
| miRNA-29a | Anti-fibrogenic | Inhibition of PIK3R3 | Fu et al. (179) |
| miRNA-29b | Anti-fibrogenic | Inhibition of STAT3 | Gong et al. (185) |
| miRNA-29b | Anti-fibrogenic | Inhibition of SMAD3 | Wang et al. (186) |
| miRNA-29b | Anti-fibrogenic | Inhibition of SMAD3 | Liang et al. (187) |
| miRNA-29b | Anti-fibrogenic | Inhibition of CTGF | Guo et al. (198) |
| miRNA-29b | Anti-fibrogenic | Inhibition of CTGF | Li et al. (199) |
| miRNA-29b | Anti-fibrogenic | Inhibition of PTHLH | Dou et al. (205) |
| miRNA-29b | Anti-fibrogenic | Inhibition of PTHLH | He et al. (208) |
| miRNA-29b | Anti-fibrogenic | Inhibition of DNMT3B | Xie et al. (211) |
| miRNA-29 | Anti-fibrogenic | Inhibition of PDGFB | Targetscan 7.2 (190) |
| miRNA-29 | Anti-fibrogenic | Inhibition of PDGFC and IGF1 | Kwiecinski et al. (175) |
| miRNA-223 | Anti-fibrogenic | Inhibition of ATG7 | Wang et al. (216) |
| miRNA-223 | Anti-fibrogenic | Inhibition of TAZ/WWTR1 | Hou et al. (54) |
| miRNA-223 | Anti-fibrogenic | Inhibition of TAZ/WWTR1 | He et al. (221) |
| miRNA-223 | Anti-fibrogenic | Inhibition of NLRP3 | Jimenez Calvente et al. (222) |
| miRNA-125b | Anti-fibrogenic | Inhibition of SMO and Hedgehog | Hyun et al. (229) |
| miRNA-125b | Anti-fibrogenic | Inhibition of SMAD2 and SMAD4 | Zhou et al. (230) |
| miRNA-125b | Fibrogenic | Inhibition of STARD13 | You et al. (228) |
| miRNA-155 | Anti-fibrogenic | Reduced EMT and ERK signaling | Dai et al. (236) |
| miRNA-155 | Fibrogenic | Inhibition of SOCS1 | Liu et al. (234) |
| miRNA-155 | Fibrogenic | HSC activation | Csak et al. (233) |
| miRNA-30a | Anti-fibrogenic | Inhibition of BECN1 | Chen et al. (244) |
| miRNA-30a | Anti-fibrogenic | Inhibition of BMP9 | Targetscan 7.2 (245) |
| miRNA-34a | Anti-fibrogenic | Inhibition of SMAD4 | Feili et al. (251) |
| miRNA-34a | Fibrogenic | Suppression of PPARγ | Li et al. (248) |
| miRNA-34a | Fibrogenic | Inhibition of ACSL1 | Yan et al. (249) |
| miRNA-34a | Fibrogenic | Sirt1-mediated HSC activation | Liu et al. (252) |
| miRNA-21 | Fibrogenic | Inhibition of PTEN | Wei et al. (257) |
| miRNA-21 | Fibrogenic | Increased PI3K-AKT signaling | Shen et al. (268) |
| miRNA-21 | Fibrogenic | Inhibition of PDCD4 | Zhang et al. (258) |
| miRNA-21 | Fibrogenic | Inhibition of HNF4A and SPRY2 | Zhao et al. (259) |
| miRNA-21 | Fibrogenic | SPRY2 and SMAD7 | Zhang et al. (261) |
| miRNA-21 | Fibrogenic | Inhibition of SRRY1 and SMAD7 | Ning et al. (264) |
Gut-liver axis
The gut-liver axis refers to the bidirectional relationship between the gut and its microbiota, and the liver, resulting from the integration of signals generated by dietary, genetic and environmental factors (274,275). Cirrhosis by itself is associated with profound alterations in gut microbiota and damage at the different levels of defense of the intestinal barrier, including the epithelial, vascular and immune barriers (274,275). The deterioration of intestinal barrier integrity and the consulting increased intestinal permeability in cirrhotic patients play a pivotal pathophysiological role in the development of severe complications (276,277). Bacterial translocation is the migration of viable or nonviable microorganisms or their pathogen-associated molecular pattern (PAMPs), such as LPS, from the gut lumen to the mesenteric lymph nodes, systemic circulation and other normally sterile extraintestinal sites (276). Via portal circulation, the liver is the first organ in the body to encounter not only absorbed nutrients, but also gut-derived bacteria and PAMPs. Chronic exposure to increased levels of PAMPs has been linked to disease progression during early stages and to infectious complications during late stages of liver disease (cirrhosis) (277). There are indications for intestinal epithelial barrier dysfunction in patients with chronic liver diseases and especially in patients with cirrhosis, which can be caused by various factors affecting both the small and large intestine (278,279). It has been demonstrated that human liver cirrhosis induces significant alterations in enterocytes’ tight junctions (277). Patients with decompensated and compensated cirrhosis presented significantly reduced expression of occludin and claudin-1 as compared to controls (280).
A shortage of autochthonous non-pathogenic bacteria and an overgrowth of potentially pathogenic bacteria are common findings in cirrhotic patients. The ratio of the amounts of beneficial autochthonous taxa (Lachnospiraceae + Ruminococaceae + Veillonellaceae + Clostridiales Incertae Sedis XIV) to those of potentially pathogenic taxa (Enterobacteriaceae + Bacteroidaceae) was low in those with early death and organ failure (281). Recently, increased levels of systemic LPS-positive bacterial EVs in patients with intestinal barrier dysfunction have been detected (282). Apparently, intestinal barrier dysfunction opens the door for bacterial EV-associated LPS that may trigger TLR4 activation of HSCs. Thus, emerging therapeutic strategies targeted at restoring intestinal eubiosis, augmenting gut barrier function and ameliorating the mucosal and systemic immune deficits that characterize and define the course of decompensated cirrhosis are in the focus of recent research (283).
There is accumulating evidence from experimental models of colitis and necrotizing enterocolitis that human, bovine, yak, porcine and rat MEX and EVs improve intestinal mucin synthesis, up-regulate the expression of tight junction proteins improving intestinal barrier function (58-61,164,165,177,258,284-293), recently reviewed elsewhere (62). In a murine model of ulcerative colitis (kindlin2 KO mice), Stremmel et al. (61) observed strong anti-inflammatory effects of orally given BMEX. In accordance, BMEX prevented colon shortening, reduced intestinal epithelium disruption, inhibited infiltration of inflammatory cells and intestinal tissue fibrosis (63). Mechanistically, BMEX attenuate inflammatory responses via inhibiting TLR4-NF-κB signaling and NLRP3 inflammasome activation (63).
Furthermore, the disturbed gut microbiota in dextran sulfate sodium-induced colitis recovered partially upon treatment with bovine MEX (63). In addition, Yu et al. (294) showed that BMEX modify bacterial growth and could promote the growth of Escherichia coli K-12 MG1655 and Lactobacillus plantarum WCFS1. Zhou et al. (295) studied the influence of dietary BMEX on bacterial communities in C57BL/6 mice. At the OTU level, four OTUs from the family of Lachnospiraceae were more than two times more abundant in mice fed BMEX compared with mice fed BMEX-deficient diet at age 7 and 47 wk. The authors concluded that BMEX modify the gut microbiome across species boundaries (295).
BMEX co-administration with anti-fibrogenic drugs
BMEX-mediated anti-fibrogenic miRNA signaling may be augmented by co-administration of drugs that further enhance anti-fibrogenic miRNA signaling such as salvianolic acid (Sal B) (112) and metformin (296). In mice, metformin could mitigate CCl4-induced HF, beneficial effects which might result from suppressed TGF-β1/SMAD3 signaling (296). Metformin directly targets and suppresses the expression of miRNA-21 (297-304), a major oncogenic and fibrogenic miRNA and component of BMEX. In C57/BL6 mice, long-term treatment with metformin reduced hepatic p-STAT3 and miRNA-21 levels and protected mice from aging-dependent hepatic vesicular steatosis (305). Importantly, metformin down-regulates the expression of lncRNA-H19, a sponge of anti-fibrotic miRNAs including miRNA-148a (98,306), let-7 (307-309), and miRNA-29b (310), respectively. Recent evidence underlines that the H19/miRNA-148a/USP4 axis facilitates HF by enhancing TGF-β signaling in both HSCs and hepatocytes (311). In fibrotic liver tissue of CCl4-induced mice, H19 over-expression significantly activated HSCs and EMT of hepatocyte both by stimulating the TGF-β pathway. H19 via sponging miRNA-148a sustained the level of ubiquitin-specific protease 4 (USP4), a direct target of miRNA-148a (312,313) stabilizing TGF-β receptor 1 signaling (311) (Figure 2).
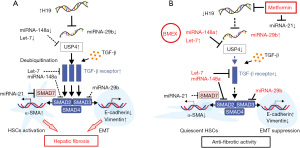
It has been proposed that metformin via AMPK activation may also inhibit the expression or activity of Lin28/Lin28A proteins, which act post-transcriptionally to decrease the levels of all let-7 family members (314-317). In a rat model of polycystic liver disease, metformin, an indirect AMPK activator and suppressor of mechanistic target of rapamycin complex 1 (mTORC1) (318), reduced liver cyst formation and HF (319). Metformin showed also preventive effects on HCC development in animal models and diabetic patients (320,321). Thus, the combination of BMEX with metformin may not only have synergistic effects in the treatment of HF but might reduce the risk of oncogenic BMEX-derived miRNAs involved in the hepatocellular cancerogenesis. Due to the fact that HF and cirrhosis enhance the risk of HCC (322), HF treatment with BMEX prior to the occurrence of HCC may decrease the risk of HCC development. Conversely, there is a risk of pre-existing hidden HCC and HCC progression that may be promoted by BMEX administration as shown in a recent rodent tumor model (73). In addition, epidemiological and translational evidence identified cow milk consumption as a nutritional risk factor of HCC (323). However, in contrast to the cancerogenic effects of the complex system milk, BMEX-derived miRNA-148a via targeting CCK-BR and USP4 may reduce the susceptibility for the development of HCC (311,324).
Thus, a combined treatment with BMEX and metformin may prevent both HF and HCC development.
Conclusions
Recent research interest focuses on siRNA- and miRNA-based therapeutics for HF (325). MiRNAs let-7b, let-7a, let-7c, miRNA-148a, let-7f, miRNA-29c, miRNA-30d, miRNA-30a and miRNA-125b, miRNA-29a and let-7d belong to the top 50 miRNAs expressed in whey-derived BMEX (211). The great majority of these miRNAs exerts anti-fibrotic effects (Table 2). The net signaling effect of all MEX-derived miRNAs should be anti-fibrogenic, because fibrogenic signaling during postnatal milk intake and the development of liver fibrosis would restrict appropriate liver growth during early life. The majority of dominant miRNAs delivered by BMEX such as miRNA-148a, let-7 and miRNA-29s are key anti-fibrotic miRNAs that inhibit HSC activation. However, there is concern about fibrogenic and oncogenic miRNAs transferred via BMEX such as miRNA-21, which could be silenced by specific RNAs loaded to BMEX prior to delivery (74,326) or co-administration of miRNA-21-suppressing drugs such as metformin (297-304).
Experimental evidence indicates that BMEX and their miRNA cargo reach the liver (70,73,75), and presumably the space of Disse including HSCs. Recent bioinformatic evidence supports the view that bovine miRNAs are able to interact with human genes (327). Future studies in rodent systems as well as HSC cell cultures exposed to BMEX are required to obtain further experimental support. The stabilization of intestinal barrier function by BMEX, the anti-inflammatory effects of BMEX and the modification of the microbiome by BMEX may be additional beneficial effects for the treatment of patients with HF. Thus, BMEX may have a direct impact on HSC homeostasis and the gut-liver axis, which is in accordance with recent views promotes the therapeutic use of BMEX per se or for drug delivery (74-77,328,329).
There is accumulating evidence that exosomes and their cargo derived from MSCs and NK cells are effective inhibitors of HSC activation and fibrogenesis (42,81,195,330). Our predictions of BMEX as potential treatment option of HF is still limited by the fact that experimental and clinical studies proving beneficial therapeutic effects of BMEX in HSC cultures, animal models of HF and in patients with HF are still missing. However, it has recently been shown in a rat model of cardiac fibrosis that administration of BMEX over a period of 7 days alleviated cardiac fibrosis and reduced the over-expression of COL1 and α-SMA (331). Exosome-encapsulated miRNA-29 decreased renal fibrosis, TGF-β, α-SMA, fibronectin, and COL1A1 in the kidney of unilateral ureteral obstruction mice (332). Accumulated basic research evidence presented in this narrative review should thus stimulate future research work to elucidate the therapeutic efficacy of oral BMEX administration for the treatment of HF.
Taken together, basic research evidence presented in this narrative review identifies BMEX as promising therapeutic agents for the treatment of HF either native or supplemented with siRNAs (antagomiR-21) or co-administered with other anti-fibrotic drugs that modify miRNA expression in HSCs.
Acknowledgments
The authors thank Sabine Weiskirchen for the creation of supportive art work.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Digestive Medicine Research for the series “The Pathogenesis of Hepatic Fibrosis: Basic Facts and Clinical Challenges”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist (available at https://dmr.amegroups.com/article/view/10.21037/dmr-21-79/rc).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dmr.amegroups.com/article/view/10.21037/dmr-21-79/coif). The series “The Pathogenesis of Hepatic Fibrosis: Basic Facts and Clinical Challenges” was commissioned by the editorial office without any funding or sponsorship. RW served as an unpaid Guest Editor of the series and serves as an unpaid editorial board member of Digestive Medicine Research from January 2022 to December 2024. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Roehlen N, Crouchet E, Baumert TF. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells 2020;9:875. [Crossref] [PubMed]
- Asrani SK, Devarbhavi H, Eaton J, et al. Burden of liver diseases in the world. J Hepatol 2019;70:151-71. [Crossref] [PubMed]
- D'Amico G, Morabito A, D'Amico M, et al. Clinical states of cirrhosis and competing risks. J Hepatol 2018;68:563-76. [Crossref] [PubMed]
- Marcellin P, Kutala BK. Liver diseases: A major, neglected global public health problem requiring urgent actions and large-scale screening. Liver Int 2018;38:2-6. [Crossref] [PubMed]
- Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest 2007;117:539-48. [Crossref] [PubMed]
- Elpek GÖ. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J Gastroenterol 2014;20:7260-76. [Crossref] [PubMed]
- Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol 2014;20:7312-24. [Crossref] [PubMed]
- Mederacke I, Hsu CC, Troeger JS, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun 2013;4:2823. [Crossref] [PubMed]
- Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev 2017;121:27-42. [Crossref] [PubMed]
- Dou C, Liu Z, Tu K, et al. P300 Acetyltransferase Mediates Stiffness-Induced Activation of Hepatic Stellate Cells Into Tumor-Promoting Myofibroblasts. Gastroenterology 2018;154:2209-2221.e14. [Crossref] [PubMed]
- Brandon-Warner E, Benbow JH, Swet JH, et al. Adeno-Associated Virus Serotype 2 Vector-Mediated Reintroduction of microRNA-19b Attenuates Hepatic Fibrosis. Hum Gene Ther 2018;29:674-86. [Crossref] [PubMed]
- Campana L, Iredale JP. Regression of Liver Fibrosis. Semin Liver Dis 2017;37:1-10. [Crossref] [PubMed]
- Karlmark KR, Weiskirchen R, Zimmermann HW, et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 2009;50:261-74. [Crossref] [PubMed]
- Ramachandran P, Pellicoro A, Vernon MA, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci U S A 2012;109:E3186-95. [Crossref] [PubMed]
- Natarajan V, Harris EN, Kidambi S. SECs (Sinusoidal Endothelial Cells), Liver Microenvironment, and Fibrosis. Biomed Res Int 2017;2017:4097205. [Crossref] [PubMed]
- Weiskirchen R, Tacke F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg Nutr 2014;3:344-63. [PubMed]
- Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017;17:306-21. [Crossref] [PubMed]
- Barron L, Wynn TA. Fibrosis is regulated by Th2 and Th17 responses and by dynamic interactions between fibroblasts and macrophages. Am J Physiol Gastrointest Liver Physiol 2011;300:G723-8. [Crossref] [PubMed]
- Weiskirchen R, Weiskirchen S, Tacke F. Organ and tissue fibrosis: Molecular signals, cellular mechanisms and translational implications. Mol Aspects Med 2019;65:2-15. [Crossref] [PubMed]
- Tacke F, Zimmermann HW. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 2014;60:1090-6. [Crossref] [PubMed]
- Fallowfield JA, Mizuno M, Kendall TJ, et al. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol 2007;178:5288-95. [Crossref] [PubMed]
- Borkham-Kamphorst E, Weiskirchen R. The PDGF system and its antagonists in liver fibrosis. Cytokine Growth Factor Rev 2016;28:53-61. [Crossref] [PubMed]
- Ying HZ, Chen Q, Zhang WY, et al. PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics Mol Med Rep 2017;16:7879-89. (Review). [Crossref] [PubMed]
- Xu F, Liu C, Zhou D, et al. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J Histochem Cytochem 2016;64:157-67. [Crossref] [PubMed]
- Nishikawa K, Osawa Y, Kimura K. Wnt/β-Catenin Signaling as a Potential Target for the Treatment of Liver Cirrhosis Using Antifibrotic Drugs. Int J Mol Sci 2018;19:3103. [Crossref] [PubMed]
- van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213-28. [Crossref] [PubMed]
- He C, Zheng S, Luo Y, et al. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018;8:237-55. [Crossref] [PubMed]
- Charrier A, Chen R, Chen L, et al. Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery 2014;156:548-55. [Crossref] [PubMed]
- Chen L, Chen R, Kemper S, et al. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1. Am J Physiol Gastrointest Liver Physiol 2015;309:G491-9. [Crossref] [PubMed]
- Seo W, Eun HS, Kim SY, et al. Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by γδ T cells in liver fibrosis. Hepatology 2016;64:616-31. [Crossref] [PubMed]
- Devhare PB, Sasaki R, Shrivastava S, et al. Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J Virol 2017;91:e02225-16. [PubMed]
- Shen J, Huang CK, Yu H, et al. The role of exosomes in hepatitis, liver cirrhosis and hepatocellular carcinoma. J Cell Mol Med 2017;21:986-92. [Crossref] [PubMed]
- Lee YS, Kim SY, Ko E, et al. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci Rep 2017;7:3710. [Crossref] [PubMed]
- Li X, Liu R, Huang Z, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 2018;68:599-615. [Crossref] [PubMed]
- Wan L, Xia T, Du Y, et al. Exosomes from activated hepatic stellate cells contain GLUT1 and PKM2: a role for exosomes in metabolic switch of liver nonparenchymal cells. FASEB J 2019;33:8530-42. [Crossref] [PubMed]
- Liu R, Li X, Zhu W, et al. Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis. Hepatology 2019;70:1317-35. [Crossref] [PubMed]
- Xiao Y, Liu R, Li X, et al. Long Noncoding RNA H19 Contributes to Cholangiocyte Proliferation and Cholestatic Liver Fibrosis in Biliary Atresia. Hepatology 2019;70:1658-73. [Crossref] [PubMed]
- Chen L, Yao X, Yao H, et al. Exosomal miR-103-3p from LPS-activated THP-1 macrophage contributes to the activation of hepatic stellate cells. FASEB J 2020;34:5178-92. [Crossref] [PubMed]
- Zhang XW, Zhou JC, Peng D, et al. Disrupting the TRIB3-SQSTM1 interaction reduces liver fibrosis by restoring autophagy and suppressing exosome-mediated HSC activation. Autophagy 2020;16:782-96. [Crossref] [PubMed]
- Shen M, Shen Y, Fan X, et al. Roles of Macrophages and Exosomes in Liver Diseases. Front Med (Lausanne) 2020;7:583691. [Crossref] [PubMed]
- Gao J, Wei B, de Assuncao TM, et al. Hepatic stellate cell autophagy inhibits extracellular vesicle release to attenuate liver fibrosis. J Hepatol 2020;73:1144-54. [Crossref] [PubMed]
- Li T, Yan Y, Wang B, et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev 2013;22:845-54. [Crossref] [PubMed]
- Cai S, Cheng X, Pan X, et al. Emerging role of exosomes in liver physiology and pathology. Hepatol Res 2017;47:194-203. [Crossref] [PubMed]
- Lou G, Chen Z, Zheng M, et al. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp Mol Med 2017;49:e346. [Crossref] [PubMed]
- Qu Y, Zhang Q, Cai X, et al. Exosomes derived from miR-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med 2017;21:2491-502. [Crossref] [PubMed]
- Ferguson SW, Wang J, Lee CJ, et al. The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci Rep 2018;8:1419. [Crossref] [PubMed]
- Chen L, Chen R, Kemper S, et al. Therapeutic effects of serum extracellular vesicles in liver fibrosis. J Extracell Vesicles 2018;7:1461505. [Crossref] [PubMed]
- Rong X, Liu J, Yao X, et al. Human bone marrow mesenchymal stem cells-derived exosomes alleviate liver fibrosis through the Wnt/β-catenin pathway. Stem Cell Res Ther 2019;10:98. [Crossref] [PubMed]
- Li X, Chen R, Kemper S, et al. Extracellular Vesicles From Hepatocytes Are Therapeutic for Toxin-Mediated Fibrosis and Gene Expression in the Liver. Front Cell Dev Biol 2020;7:368. [Crossref] [PubMed]
- Bruno S, Chiabotto G, Camussi G. Extracellular Vesicles: A Therapeutic Option for Liver Fibrosis. Int J Mol Sci 2020;21:4255. [Crossref] [PubMed]
- Zhu M, Liu X, Li W, et al. Exosomes derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis via activating autophagy. Hum Exp Toxicol 2020;39:1619-27. [Crossref] [PubMed]
- Wang L, Wang Y, Quan J. Exosomes derived from natural killer cells inhibit hepatic stellate cell activation and liver fibrosis. Hum Cell 2020;33:582-9. [Crossref] [PubMed]
- Chiabotto G, Pasquino C, Camussi G, et al. Molecular Pathways Modulated by Mesenchymal Stromal Cells and Their Extracellular Vesicles in Experimental Models of Liver Fibrosis. Front Cell Dev Biol 2020;8:594794. [Crossref] [PubMed]
- Hou X, Yin S, Ren R, et al. Myeloid-Cell-Specific IL-6 Signaling Promotes MicroRNA-223-Enriched Exosome Production to Attenuate NAFLD-Associated Fibrosis. Hepatology 2021;74:116-32. [Crossref] [PubMed]
- Brigstock DR. Extracellular Vesicles in Organ Fibrosis: Mechanisms, Therapies, and Diagnostics. Cells 2021;10:1596. [Crossref] [PubMed]
- Ding J, Wang J, Chen J. Exosomes as therapeutic vehicles in liver diseases. Ann Transl Med 2021;9:735. [Crossref] [PubMed]
- Schuppan D, Ashfaq-Khan M, Yang AT, et al. Liver fibrosis: Direct antifibrotic agents and targeted therapies. Matrix Biol 2018;68-69:435-51. [Crossref] [PubMed]
- Benmoussa A, Diallo I, Salem M, et al. Concentrates of two subsets of extracellular vesicles from cow's milk modulate symptoms and inflammation in experimental colitis. Sci Rep 2019;9:14661. [Crossref] [PubMed]
- Chen W, Wang X, Yan X, et al. The emerging role of exosomes in the pathogenesis, prognosis and treatment of necrotizing enterocolitis. Am J Transl Res 2020;12:7020-33. [PubMed]
- Reif S, Elbaum-Shiff Y, Koroukhov N, et al. Cow and Human Milk-Derived Exosomes Ameliorate Colitis in DSS Murine Model. Nutrients 2020;12:2589. [Crossref] [PubMed]
- Stremmel W, Weiskirchen R, Melnik BC. Milk Exosomes Prevent Intestinal Inflammation in a Genetic Mouse Model of Ulcerative Colitis: A Pilot Experiment. Inflamm Intest Dis 2020;5:117-23. [Crossref] [PubMed]
- Melnik BC, Stremmel W, Weiskirchen R, et al. Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development. Biomolecules 2021;11:851. [Crossref] [PubMed]
- Tong L, Hao H, Zhang Z, et al. Milk-derived extracellular vesicles alleviate ulcerative colitis by regulating the gut immunity and reshaping the gut microbiota. Theranostics 2021;11:8570-86. [Crossref] [PubMed]
- Baier SR, Nguyen C, Xie F, et al. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 2014;144:1495-500. [Crossref] [PubMed]
- Melnik BC, John SM, Schmitz G. Milk: an exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J Transl Med 2014;12:43. [Crossref] [PubMed]
- Arntz OJ, Pieters BC, Oliveira MC, et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol Nutr Food Res 2015;59:1701-12. [Crossref] [PubMed]
- Melnik BC, Schmitz G. MicroRNAs: Milk's epigenetic regulators. Best Pract Res Clin Endocrinol Metab 2017;31:427-42. [Crossref] [PubMed]
- Wang L, Sadri M, Giraud D, et al. RNase H2-Dependent Polymerase Chain Reaction and Elimination of Confounders in Sample Collection, Storage, and Analysis Strengthen Evidence That microRNAs in Bovine Milk Are Bioavailable in Humans. J Nutr 2018;148:153-9. [Crossref] [PubMed]
- Kusuma RJ, Manca S, Friemel T, et al. Human vascular endothelial cells transport foreign exosomes from cow's milk by endocytosis. Am J Physiol Cell Physiol 2016;310:C800-7. [Crossref] [PubMed]
- Manca S, Upadhyaya B, Mutai E, et al. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep 2018;8:11321. [Crossref] [PubMed]
- Benmoussa A, Provost P. Milk MicroRNAs in Health and Disease. Compr Rev Food Sci Food Saf 2019;18:703-22. [Crossref] [PubMed]
- Sadri M, Shu J, Kachman SD, et al. Milk exosomes and miRNA cross the placenta and promote embryo survival in mice. Reproduction 2020;160:501-9. [Crossref] [PubMed]
- Samuel M, Fonseka P, Sanwlani R, et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat Commun 2021;12:3950. [Crossref] [PubMed]
- Munagala R, Aqil F, Jeyabalan J, et al. Bovine milk-derived exosomes for drug delivery. Cancer Lett 2016;371:48-61. [Crossref] [PubMed]
- Betker JL, Angle BM, Graner MW, et al. The Potential of Exosomes From Cow Milk for Oral Delivery. J Pharm Sci 2019;108:1496-505. [Crossref] [PubMed]
- Sanwlani R, Fonseka P, Chitti SV, et al. Milk-Derived Extracellular Vesicles in Inter-Organism, Cross-Species Communication and Drug Delivery. Proteomes 2020;8:11. [Crossref] [PubMed]
- Ong SL, Blenkiron C, Haines S, et al. Ruminant Milk-Derived Extracellular Vesicles: A Nutritional and Therapeutic Opportunity? Nutrients 2021;13:2505. [Crossref] [PubMed]
- Shen H, Zhang M, Kaita K, et al. Expression of Fc Fragment Receptors of Immunoglobulin G (FcγRs) in Rat Hepatic Stellate Cells. Dig Dis Sci 2005;50:181-7. [Crossref]
- Aguilar-Lozano A, Baier S, Grove R, et al. Concentrations of Purine Metabolites Are Elevated in Fluids from Adults and Infants and in Livers from Mice Fed Diets Depleted of Bovine Milk Exosomes and their RNA Cargos. J Nutr 2018;148:1886-94. [Crossref] [PubMed]
- López de Las Hazas MC, Del Pozo-Acebo L, Hansen MS, et al. Dietary bovine milk miRNAs transported in extracellular vesicles are partially stable during GI digestion, are bioavailable and reach target tissues but need a minimum dose to impact on gene expression. Eur J Nutr 2022;61:1043-56. [Crossref] [PubMed]
- An SY, Jang YJ, Lim HJ, et al. Milk Fat Globule-EGF Factor 8, Secreted by Mesenchymal Stem Cells, Protects Against Liver Fibrosis in Mice. Gastroenterology 2017;152:1174-86. [Crossref] [PubMed]
- Fujiwara C, Uehara A, Sekiguchi A, et al. Suppressive Regulation by MFG-E8 of Latent Transforming Growth Factor β-Induced Fibrosis via Binding to αv Integrin: Significance in the Pathogenesis of Fibrosis in Systemic Sclerosis. Arthritis Rheumatol 2019;71:302-14. [Crossref] [PubMed]
- Motegi S, Garfield S, Feng X, et al. Potentiation of platelet-derived growth factor receptor-β signaling mediated by integrin-associated MFG-E8. Arterioscler Thromb Vasc Biol 2011;31:2653-64. [Crossref] [PubMed]
- Reinhardt TA, Lippolis JD, Nonnecke BJ, et al. Bovine milk exosome proteome. J Proteomics 2012;75:1486-92. [Crossref] [PubMed]
- Yamauchi M, Shimizu K, Rahman M, et al. Efficient method for isolation of exosomes from raw bovine milk. Drug Dev Ind Pharm 2019;45:359-64. [Crossref] [PubMed]
- Rahman MM, Shimizu K, Yamauchi M, et al. Acidification effects on isolation of extracellular vesicles from bovine milk. PLoS One 2019;14:e0222613. [Crossref] [PubMed]
- He Y, Huang C, Zhang SP, et al. The potential of microRNAs in liver fibrosis. Cell Signal 2012;24:2268-72. [Crossref] [PubMed]
- Kitano M, Bloomston PM. Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis. J Clin Med 2016;5:38. [Crossref] [PubMed]
- Zhu S, Chen X, Chen Y, et al. Role of microRNAs in Hepatic Stellate Cells and Hepatic Fibrosis: An Update. Curr Pharm Des 2021;27:3000-11. [Crossref] [PubMed]
- Hyun J, Jung Y. MicroRNAs in liver fibrosis: Focusing on the interaction with hedgehog signaling. World J Gastroenterol 2016;22:6652-62. [Crossref] [PubMed]
- Yang JJ, Tao H, Li J. Hedgehog signaling pathway as key player in liver fibrosis: new insights and perspectives. Expert Opin Ther Targets 2014;18:1011-21. [Crossref] [PubMed]
- Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer 2013;12:43. [Crossref] [PubMed]
- Zhao Q, Qin CY, Zhao ZH, et al. Epigenetic modifications in hepatic stellate cells contribute to liver fibrosis. Tohoku J Exp Med 2013;229:35-43. [Crossref] [PubMed]
- El Taghdouini A, Sørensen AL, Reiner AH, et al. Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells. Oncotarget 2015;6:26729-45. [Crossref] [PubMed]
- El Taghdouini A, van Grunsven LA. Epigenetic regulation of hepatic stellate cell activation and liver fibrosis. Expert Rev Gastroenterol Hepatol 2016;10:1397-408. [Crossref] [PubMed]
- Barcena-Varela M, Colyn L, Fernandez-Barrena MG. Epigenetic Mechanisms in Hepatic Stellate Cell Activation During Liver Fibrosis and Carcinogenesis. Int J Mol Sci 2019;20:2507. [Crossref] [PubMed]
- Yang JJ, She Q, Yang Y, et al. DNMT1 controls LncRNA H19/ERK signal pathway in hepatic stellate cell activation and fibrosis. Toxicol Lett 2018;295:325-34. [Crossref] [PubMed]
- Zeng J, Zhu L, Liu J, et al. Metformin Protects against Oxidative Stress Injury Induced by Ischemia/Reperfusion via Regulation of the lncRNA-H19/miR-148a-3p/Rock2 Axis. Oxid Med Cell Longev 2019;2019:8768327. [Crossref] [PubMed]
- Yang JJ, Liu LP, Tao H, et al. MeCP2 silencing of LncRNA H19 controls hepatic stellate cell proliferation by targeting IGF1R. Toxicology 2016;359-360:39-46. [Crossref] [PubMed]
- Huang W, Huang F, Zhang R, et al. LncRNA Neat1 expedites the progression of liver fibrosis in mice through targeting miR-148a-3p and miR-22-3p to upregulate Cyth3. Cell Cycle 2021;20:490-507. [Crossref] [PubMed]
- Pan W, Zhu S, Yuan M, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol 2010;184:6773-81. [Crossref] [PubMed]
- Liu L, Yi H, Wang C, et al. Integrated Nanovaccine with MicroRNA-148a Inhibition Reprograms Tumor-Associated Dendritic Cells by Modulating miR-148a/DNMT1/SOCS1 Axis. J Immunol 2016;197:1231-41. [Crossref] [PubMed]
- Seki E, Brenner DA. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci 2015;22:512-8. [Crossref] [PubMed]
- Luo J, Xie M, Hou Y, et al. A novel epigenetic mechanism unravels hsa-miR-148a-3p-mediated CYP2B6 downregulation in alcoholic hepatitis disease. Biochem Pharmacol 2021;188:114582. [Crossref] [PubMed]
- Heo MJ, Kim TH, You JS, et al. Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression. Gut 2019;68:708-20. [Crossref] [PubMed]
- Luo J, Hou Y, Ma W, et al. A novel mechanism underlying alcohol dehydrogenase expression: hsa-miR-148a-3p promotes ADH4 expression via an AGO1-dependent manner in control and ethanol-exposed hepatic cells. Biochem Pharmacol 2021;189:114458. [Crossref] [PubMed]
- Tomela K, Karolak JA, Ginter-Matuszewska B, et al. Influence of TGFBR2, TGFB3, DNMT1, and DNMT3A Knockdowns on CTGF, TGFBR2, and DNMT3A in Neonatal and Adult Human Dermal Fibroblasts Cell Lines. Curr Issues Mol Biol 2021;43:276-85. [Crossref] [PubMed]
- Ramazani Y, Knops N, Elmonem MA, et al. Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol 2018;68-69:44-66. [Crossref] [PubMed]
- Seo HY, Lee SH, Lee JH, et al. Src Inhibition Attenuates Liver Fibrosis by Preventing Hepatic Stellate Cell Activation and Decreasing Connetive Tissue Growth Factor. Cells 2020;9:558. [Crossref] [PubMed]
- Gressner OA, Gressner AM. Connective tissue growth factor: a fibrogenic master switch in fibrotic liver diseases. Liver Int 2008;28:1065-79. [Crossref] [PubMed]
- Huang G, Brigstock DR. Regulation of hepatic stellate cells by connective tissue growth factor. Front Biosci (Landmark Ed) 2012;17:2495-507. [Crossref] [PubMed]
- Yu F, Lu Z, Chen B, et al. Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis by attenuating DNMT1-mediated Patched1 methylation. J Cell Mol Med 2015;19:2617-32. [Crossref] [PubMed]
- targetscan.org: DNMT1/miR-148a/miR-148b/miR-152, http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs= NST00000340748.4&taxid=9606&members=miR-148-3p/152-3p&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1
- Golan-Gerstl R, Elbaum Shiff Y, Moshayoff V, et al. Characterization and biological function of milk-derived miRNAs. Mol Nutr Food Res 2017; [Crossref] [PubMed]
- Reif S, Elbaum Shiff Y, Golan-Gerstl R. Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner. J Transl Med 2019;17:325. [Crossref] [PubMed]
- Chen X, Gao C, Li H, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res 2010;20:1128-37. [Crossref] [PubMed]
- van Herwijnen MJC, Driedonks TAP, Snoek BL, et al. Abundantly Present miRNAs in Milk-Derived Extracellular Vesicles Are Conserved Between Mammals. Front Nutr 2018;5:81. [Crossref] [PubMed]
- miRBase. Stem-loop sequence hsa-mir-148a, https://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000253
- miRBase. Stem-loop sequence bta-mir-148a, https://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0004737
- Cintio M, Polacchini G, Scarsella E, et al. MicroRNA Milk Exosomes: From Cellular Regulator to Genomic Marker. Animals (Basel) 2020;10:1126. [Crossref] [PubMed]
- Dewidar B, Meyer C, Dooley S, et al. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells 2019;8:1419. [Crossref] [PubMed]
- Derynck R, Budi EH. Specificity, versatility, and control of TGF-β family signaling. Sci Signal 2019;12:aav5183. [Crossref] [PubMed]
- Koo JH, Lee HJ, Kim W, et al. Endoplasmic Reticulum Stress in Hepatic Stellate Cells Promotes Liver Fibrosis via PERK-Mediated Degradation of HNRNPA1 and Up-regulation of SMAD2. Gastroenterology 2016;150:181-193.e8. [Crossref] [PubMed]
- Cai X, Li Z, Zhang Q, et al. CXCL6-EGFR-induced Kupffer cells secrete TGF-β1 promoting hepatic stellate cell activation via the SMAD2/BRD4/C-MYC/EZH2 pathway in liver fibrosis. J Cell Mol Med 2018;22:5050-61. [Crossref] [PubMed]
- Jiang F, Mu J, Wang X, et al. The repressive effect of miR-148a on TGF beta-SMADs signal pathway is involved in the glabridin-induced inhibition of the cancer stem cells-like properties in hepatocellular carcinoma cells. PLoS One 2014;9:e96698. [Crossref] [PubMed]
- Nadella S, Burks J, Al-Sabban A, et al. Dietary fat stimulates pancreatic cancer growth and promotes fibrosis of the tumor microenvironment through the cholecystokinin receptor. Am J Physiol Gastrointest Liver Physiol 2018;315:G699-712. [Crossref] [PubMed]
- Smith JP, Cooper TK, McGovern CO, et al. Cholecystokinin receptor antagonist halts progression of pancreatic cancer precursor lesions and fibrosis in mice. Pancreas 2014;43:1050-9. [Crossref] [PubMed]
- Tucker RD, Ciofoaia V, Nadella S, et al. A Cholecystokinin Receptor Antagonist Halts Nonalcoholic Steatohepatitis and Prevents Hepatocellular Carcinoma. Dig Dis Sci 2020;65:189-203. [Crossref] [PubMed]
- Yu B, Lv X, Su L, et al. MiR-148a Functions as a Tumor Suppressor by Targeting CCK-BR via Inactivating STAT3 and Akt in Human Gastric Cancer. PLoS One 2016;11:e0158961. [Crossref] [PubMed]
- Nariman-Saleh-Fam Z, Bastami M, Ardalan M, et al. Cell-free microRNA-148a is associated with renal allograft dysfunction: Implication for biomarker discovery. J Cell Biochem 2019;120:5737-46. [Crossref] [PubMed]
- McDaniel K, Hall C, Sato K, et al. Lin28 and let-7: roles and regulation in liver diseases. Am J Physiol Gastrointest Liver Physiol 2016;310:G757-65. [Crossref] [PubMed]
- Liu Y, Nie H, Ding Y, et al. MiRNA, a New Treatment Strategy for Pulmonary Fibrosis. Curr Drug Targets 2021;22:793-802. [Crossref] [PubMed]
- McDaniel K, Huang L, Sato K, et al. The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. J Biol Chem 2017;292:11336-47. [Crossref] [PubMed]
- Nam Y, Chen C, Gregory RI, et al. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011;147:1080-91. [Crossref] [PubMed]
- Terzuoli E, Nannelli G, Giachetti A, et al. Targeting endothelial-to-mesenchymal transition: the protective role of hydroxytyrosol sulfate metabolite. Eur J Nutr 2020;59:517-27. [Crossref] [PubMed]
- Bi WR, Yang CQ, Shi Q. Transforming growth factor-β1 induced epithelial-mesenchymal transition in hepatic fibrosis. Hepatogastroenterology 2012;59:1960-3. [PubMed]
- Shrestha N, Chand L, Han MK, et al. Glutamine inhibits CCl4 induced liver fibrosis in mice and TGF-β1 mediated epithelial-mesenchymal transition in mouse hepatocytes. Food Chem Toxicol 2016;93:129-37. [Crossref] [PubMed]
- Zhang Q, Chang X, Wang H, et al. TGF-β1 mediated Smad signaling pathway and EMT in hepatic fibrosis induced by Nano NiO in vivo and in vitro. Environ Toxicol 2020;35:419-29. [Crossref] [PubMed]
- Bronevetsky Y, Burt TD, McCune JM. Lin28b Regulates Fetal Regulatory T Cell Differentiation through Modulation of TGF-β Signaling. J Immunol 2016;197:4344-50. [Crossref] [PubMed]
- Shen GY, Ren H, Shang Q, et al. Let-7f-5p regulates TGFBR1 in glucocorticoid-inhibited osteoblast differentiation and ameliorates glucocorticoid-induced bone loss. Int J Biol Sci 2019;15:2182-97. [Crossref] [PubMed]
- Sun L, Zhu M, Feng W, et al. Exosomal miRNA Let-7 from Menstrual Blood-Derived Endometrial Stem Cells Alleviates Pulmonary Fibrosis through Regulating Mitochondrial DNA Damage. Oxid Med Cell Longev 2019;2019:4506303. [Crossref] [PubMed]
- Pieters BC, Arntz OJ, Bennink MB, et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS One 2015;10:e0121123. [Crossref] [PubMed]
- Benmoussa A, Lee CH, Laffont B, et al. Commercial Dairy Cow Milk microRNAs Resist Digestion under Simulated Gastrointestinal Tract Conditions. J Nutr 2016;146:2206-15. [Crossref] [PubMed]
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 2005;25:9198-208. [Crossref] [PubMed]
- Ahn G, Kim YH, Ahn JY. Multifaceted effects of milk-exosomes (Mi-Exo) as a modulator of scar-free wound healing. Nanoscale Adv 2021;3:528-37. [Crossref]
- Paik YH, Schwabe RF, Bataller R, et al. Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology 2003;37:1043-55. [Crossref] [PubMed]
- Pradere JP, Troeger JS, Dapito DH, et al. Toll-like receptor 4 and hepatic fibrogenesis. Semin Liver Dis 2010;30:232-44. [Crossref] [PubMed]
- Seki E, De Minicis S, Osterreicher CH, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 2007;13:1324-32. [Crossref] [PubMed]
- Guo J, Friedman SL. Toll-like receptor 4 signaling in liver injury and hepatic fibrogenesis. Fibrogenesis Tissue Repair 2010;3:21. [Crossref] [PubMed]
- Petrasek J, Csak T, Szabo G. Toll-like receptors in liver disease. Adv Clin Chem 2013;59:155-201. [Crossref] [PubMed]
- Ouyang Y, Guo J, Lin C, et al. Transcriptomic analysis of the effects of Toll-like receptor 4 and its ligands on the gene expression network of hepatic stellate cells. Fibrogenesis Tissue Repair 2016;9:2. [Crossref] [PubMed]
- Chen Y, Wu Z, Yuan B, et al. MicroRNA-146a-5p attenuates irradiation-induced and LPS-induced hepatic stellate cell activation and hepatocyte apoptosis through inhibition of TLR4 pathway. Cell Death Dis 2018;9:22. [Crossref] [PubMed]
- Liao X, Zhan W, Tian T, et al. MicroRNA-326 attenuates hepatic stellate cell activation and liver fibrosis by inhibiting TLR4 signaling. J Cell Biochem 2019; [Crossref] [PubMed]
- Lin ZP, Lin HL, Yu XP, et al. TLR4 mediates inflammation and hepatic fibrosis induced by chronic intermittent hypoxia in rats. Mol Med Rep 2020;22:651-60. [Crossref] [PubMed]
- Liu Y, Nong L, Jia Y, et al. Aspirin alleviates hepatic fibrosis by suppressing hepatic stellate cells activation via the TLR4/NF-κB pathway. Aging (Albany NY) 2020;12:6058-66. [Crossref] [PubMed]
- Li B, Yan C, Wu J, et al. Clonorchis sinensis ESPs enhance the activation of hepatic stellate cells by a cross-talk of TLR4 and TGF-β/Smads signaling pathway. Acta Trop 2020;205:105307. [Crossref] [PubMed]
- Dong Z, Zhuang Q, Ning M, et al. Palmitic acid stimulates NLRP3 inflammasome activation through TLR4-NF-κB signal pathway in hepatic stellate cells. Ann Transl Med 2020;8:168. [Crossref] [PubMed]
- Jiang K, Yang J, Yang C, et al. miR-148a suppresses inflammation in lipopolysaccharide-induced endometritis. J Cell Mol Med 2020;24:405-17. [Crossref] [PubMed]
- Liu X, Zhan Z, Xu L, et al. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J Immunol 2010;185:7244-51. [Crossref] [PubMed]
- Zheng D, He D, Lu X, et al. The miR-148a alleviates hepatic ischemia/reperfusion injury in mice via targeting CaMKIIα. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2016;32:1202-6. [PubMed]
- Zheng D, Li Z, Wei X, et al. Role of miR-148a in Mitigating Hepatic Ischemia-Reperfusion Injury by Repressing the TLR4 Signaling Pathway via Targeting CaMKIIα in Vivo and in Vitro. Cell Physiol Biochem 2018;49:2060-72. [Crossref] [PubMed]
- Patel V, Carrion K, Hollands A, et al. The stretch responsive microRNA miR-148a-3p is a novel repressor of IKBKB, NF-κB signaling, and inflammatory gene expression in human aortic valve cells. FASEB J 2015;29:1859-68. [Crossref] [PubMed]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009;139:693-706. [Crossref] [PubMed]
- Xie MY, Hou LJ, Sun JJ, et al. Porcine Milk Exosome MiRNAs Attenuate LPS-Induced Apoptosis through Inhibiting TLR4/NF-κB and p53 Pathways in Intestinal Epithelial Cells. J Agric Food Chem 2019;67:9477-91. [Crossref] [PubMed]
- Gao R, Zhang R, Qian T, et al. A comparison of exosomes derived from different periods breast milk on protecting against intestinal organoid injury. Pediatr Surg Int 2019;35:1363-8. [Crossref] [PubMed]
- Breitkopf K. Expression patterns of PDGF-A, -B, -C and -D and the PDGF-receptors alpha and beta in activated rat hepatic stellate cells (HSC). Cytokine 2005;31:349-57. [Crossref] [PubMed]
- Kocabayoglu P, Lade A, Lee YA, et al. β-PDGF receptor expressed by hepatic stellate cells regulates fibrosis in murine liver injury, but not carcinogenesis. J Hepatol 2015;63:141-7. [Crossref] [PubMed]
- Czochra P, Klopcic B, Meyer E, et al. Liver fibrosis induced by hepatic overexpression of PDGF-B in transgenic mice. J Hepatol 2006;45:419-28. [Crossref] [PubMed]
- Noetel A, Kwiecinski M, Elfimova N, et al. microRNA are Central Players in Anti- and Profibrotic Gene Regulation during Liver Fibrosis. Front Physiol 2012;3:49. [Crossref] [PubMed]
- He Y, Huang C, Lin X, et al. MicroRNA-29 family, a crucial therapeutic target for fibrosis diseases. Biochimie 2013;95:1355-9. [Crossref] [PubMed]
- Bian EB, Li J, Zhao B. miR-29, a potential therapeutic target for liver fibrosis. Gene 2014;544:259-60. [Crossref] [PubMed]
- Matsumoto Y, Itami S, Kuroda M, et al. MiR-29a Assists in Preventing the Activation of Human Stellate Cells and Promotes Recovery From Liver Fibrosis in Mice. Mol Ther 2016;24:1848-59. [Crossref] [PubMed]
- Deng Z, He Y, Yang X, et al. MicroRNA-29: A Crucial Player in Fibrotic Disease. Mol Diagn Ther 2017;21:285-94. [Crossref] [PubMed]
- Huang YH, Yang YL, Wang FS. The Role of miR-29a in the Regulation, Function, and Signaling of Liver Fibrosis. Int J Mol Sci 2018;19:1889. [Crossref] [PubMed]
- Kwiecinski M, Elfimova N, Noetel A, et al. Expression of platelet-derived growth factor-C and insulin-like growth factor I in hepatic stellate cells is inhibited by miR-29. Lab Invest 2012;92:978-87. [Crossref] [PubMed]
- Sanz S, Pucilowska JB, Liu S, et al. Expression of insulin-like growth factor I by activated hepatic stellate cells reduces fibrogenesis and enhances regeneration after liver injury. Gut 2005;54:134-41. [Crossref] [PubMed]
- Li B, Hock A, Wu RY, et al. Bovine milk-derived exosomes enhance goblet cell activity and prevent the development of experimental necrotizing enterocolitis. PLoS One 2019;14:e0211431. [Crossref] [PubMed]
- Argon Y, Bresson SE, Marzec MT, et al. Glucose-Regulated Protein 94 (GRP94): A Novel Regulator of Insulin-Like Growth Factor Production. Cells 2020;9:1844. [Crossref] [PubMed]
- Fu J, Wu B, Zhong S, et al. miR-29a-3p suppresses hepatic fibrosis pathogenesis by modulating hepatic stellate cell proliferation via targeting PIK3R3 gene expression. Biochem Biophys Res Commun 2020;529:922-9. [Crossref] [PubMed]
- Yang YL, Wang FS, Li SC, et al. MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. Int J Mol Sci 2017;18:192. [Crossref] [PubMed]
- Yang YL, Kuo HC, Wang FS, et al. MicroRNA-29a Disrupts DNMT3b to Ameliorate Diet-Induced Non-Alcoholic Steatohepatitis in Mice. Int J Mol Sci 2019;20:1499. [Crossref] [PubMed]
- Mannaerts I, Eysackers N, Onyema OO, et al. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS One 2013;8:e55786. [Crossref] [PubMed]
- Huang YH, Tiao MM, Huang LT, et al. Activation of Mir-29a in Activated Hepatic Stellate Cells Modulates Its Profibrogenic Phenotype through Inhibition of Histone Deacetylases 4. PLoS One 2015;10:e0136453. [Crossref] [PubMed]
- Lin HY, Wang FS, Yang YL, et al. MicroRNA-29a Suppresses CD36 to Ameliorate High Fat Diet-Induced Steatohepatitis and Liver Fibrosis in Mice. Cells 2019;8:1298. [Crossref] [PubMed]
- Gong X, Wang X, Zhou F. Liver microRNA-29b-3p positively correlates with relative enhancement values of magnetic resonance imaging and represses liver fibrosis. J Biochem 2020;168:603-9. [Crossref] [PubMed]
- Wang J, Chu ES, Chen HY, et al. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015;6:7325-38. [Crossref] [PubMed]
- Liang C, Bu S, Fan X. Suppressive effect of microRNA-29b on hepatic stellate cell activation and its crosstalk with TGF-β1/Smad3. Cell Biochem Funct 2016;34:326-33. [Crossref] [PubMed]
- Yu F, Chen B, Dong P, et al. HOTAIR Epigenetically Modulates PTEN Expression via MicroRNA-29b: A Novel Mechanism in Regulation of Liver Fibrosis. Mol Ther 2017;25:205-17. [Crossref] [PubMed]
- Sekiya Y, Ogawa T, Yoshizato K, et al. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem Biophys Res Commun 2011;412:74-9. [Crossref] [PubMed]
- Targetscan 7.2. Human PDGFB/miRNA-29b-3p, http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs= ENST00000331163.6&taxid=9606&showcnc=0&shownc=0&shownc_nc= &showncf1=&showncf2=&subset=1
- Targetscan. Bovine PDGFB-miRNA-29b-3p, http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs= ENST00000331163.6&taxid=9913&members=miR-29-3p&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1
- Targetscan. Human PDGFB-miRNA-29b-3p, http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000261799.4&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1
- Wu J, Huang J, Kuang S, et al. Synergistic MicroRNA Therapy in Liver Fibrotic Rat Using MRI-Visible Nanocarrier Targeting Hepatic Stellate Cells. Adv Sci (Weinh) 2019;6:1801809. [Crossref] [PubMed]
- Park HJ, Kim HG, Wang JH, et al. Comparison of TGF-β, PDGF, and CTGF in hepatic fibrosis models using DMN, CCl4, and TAA. Drug Chem Toxicol 2016;39:111-8. [PubMed]
- Chen L, Chen R, Velazquez VM, et al. Fibrogenic Signaling Is Suppressed in Hepatic Stellate Cells through Targeting of Connective Tissue Growth Factor (CCN2) by Cellular or Exosomal MicroRNA-199a-5p. Am J Pathol 2016;186:2921-33. [Crossref] [PubMed]
- Chen L, Charrier A, Zhou Y, et al. Epigenetic regulation of connective tissue growth factor by MicroRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology 2014;59:1118-29. [Crossref] [PubMed]
- Wu D, Lu P, Mi X, et al. Exosomal miR-214 from endometrial stromal cells inhibits endometriosis fibrosis. Mol Hum Reprod 2018;24:357-65. [Crossref] [PubMed]
- Guo J, Lin Q, Shao Y, et al. miR-29b promotes skin wound healing and reduces excessive scar formation by inhibition of the TGF-β1/Smad/CTGF signaling pathway. Can J Physiol Pharmacol 2017;95:437-42. [Crossref] [PubMed]
- Li J, Du S, Sheng X, et al. MicroRNA-29b Inhibits Endometrial Fibrosis by Regulating the Sp1-TGF-β1/Smad-CTGF Axis in a Rat Model. Reprod Sci 2016;23:386-94. [Crossref] [PubMed]
- Aoyama E, Kubota S, Takigawa M. CCN2/CTGF binds to fibroblast growth factor receptor 2 and modulates its signaling. FEBS Lett 2012;586:4270-5. [Crossref] [PubMed]
- Steiling H, Mühlbauer M, Bataille F, et al. Activated hepatic stellate cells express keratinocyte growth factor in chronic liver disease. Am J Pathol 2004;165:1233-41. [Crossref] [PubMed]
- Bertrand-Philippe M, Ruddell RG, Arthur MJ, et al. Regulation of tissue inhibitor of metalloproteinase 1 gene transcription by RUNX1 and RUNX2. J Biol Chem 2004;279:24530-9. [Crossref] [PubMed]
- Hattori S, Dhar DK, Hara N, et al. FR-167653, a selective p38 MAPK inhibitor, exerts salutary effect on liver cirrhosis through downregulation of Runx2. Lab Invest 2007;87:591-601. [Crossref] [PubMed]
- Li Z, Hassan MQ, Jafferji M, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 2009;284:15676-84. [Crossref] [PubMed]
- Dou P, He Y, Yu B, et al. Downregulation of microRNA-29b by DNMT3B decelerates chondrocyte apoptosis and the progression of osteoarthritis via PTHLH/CDK4/RUNX2 axis. Aging (Albany NY) 2020;13:7676-90. [Crossref] [PubMed]
- Fabrega E, Rivero M, Pons-Romero F, et al. Parathyroid hormone-related protein in liver cirrhosis. Dig Dis Sci 2000;45:703. [Crossref] [PubMed]
- Liang FF, Liu CP, Li LX, et al. Activated effects of parathyroid hormone-related protein on human hepatic stellate cells. PLoS One 2013;8:e76517. [Crossref] [PubMed]
- He S, Tang J, Diao N, et al. Parathyroid hormone-related protein activates HSCs via hedgehog signalling during liver fibrosis development. Artif Cells Nanomed Biotechnol 2019;47:1984-94. [Crossref] [PubMed]
- He S, Xue M, Liu C, et al. Parathyroid Hormone-Like Hormone Induces Epithelial-to-Mesenchymal Transition of Intestinal Epithelial Cells by Activating the Runt-Related Transcription Factor 2. Am J Pathol 2018;188:1374-88. [Crossref] [PubMed]
- Sun M, Tan L, Hu M. The role of autophagy in hepatic fibrosis. Am J Transl Res 2021;13:5747-57. [PubMed]
- Xie Z, Wu Y, Liu S, et al. LncRNA-SNHG7/miR-29b/DNMT3A axis affects activation, autophagy and proliferation of hepatic stellate cells in liver fibrosis. Clin Res Hepatol Gastroenterol 2021;45:101469. [Crossref] [PubMed]
- Howard KM, Jati Kusuma R, Baier SR, et al. Loss of miRNAs during processing and storage of cow's (Bos taurus) milk. J Agric Food Chem 2015;63:588-92. [Crossref] [PubMed]
- Izumi H, Tsuda M, Sato Y, et al. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J Dairy Sci 2015;98:2920-33. [Crossref] [PubMed]
- Yu S, Zhao Z, Sun L, et al. Fermentation Results in Quantitative Changes in Milk-Derived Exosomes and Different Effects on Cell Growth and Survival. J Agric Food Chem 2017;65:1220-8. [Crossref] [PubMed]
- Wang X, Zheng Z, Caviglia JM, et al. Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab 2016;24:848-62. [Crossref] [PubMed]
- Wang L, Wang Y, Quan J. Exosomal miR-223 derived from natural killer cells inhibits hepatic stellate cell activation by suppressing autophagy. Mol Med 2020;26:81. [Crossref] [PubMed]
- Yu K, Li N, Cheng Q, et al. miR-96-5p prevents hepatic stellate cell activation by inhibiting autophagy via ATG7. J Mol Med (Berl) 2018;96:65-74. [Crossref] [PubMed]
- Hernández-Gea V, Ghiassi-Nejad Z, Rozenfeld R, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 2012;142:938-46. [Crossref] [PubMed]
- Guillot A, Tacke F. The Unexpected Role of Neutrophils for Resolving Liver Inflammation by Transmitting MicroRNA-223 to Macrophages. Hepatology 2020;71:749-51. [Crossref] [PubMed]
- Calvente CJ, Tameda M, Johnson CD, et al. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J Clin Invest 2019;129:4091-109. [Crossref] [PubMed]
- He Y, Hwang S, Cai Y, et al. MicroRNA-223 Ameliorates Nonalcoholic Steatohepatitis and Cancer by Targeting Multiple Inflammatory and Oncogenic Genes in Hepatocytes. Hepatology 2019;70:1150-67. [Crossref] [PubMed]
- Jimenez Calvente C, Del Pilar H, Tameda M, et al. MicroRNA 223 3p Negatively Regulates the NLRP3 Inflammasome in Acute and Chronic Liver Injury. Mol Ther 2020;28:653-63. [Crossref] [PubMed]
- Liu W, Feng R, Li X, et al. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging (Albany NY) 2019;11:9569-80. [Crossref] [PubMed]
- Guan X, Gao Y, Zhou J, et al. miR-223 Regulates Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells Through a C/EBPs/miR-223/FGFR2 Regulatory Feedback Loop. Stem Cells 2015;33:1589-600. [Crossref] [PubMed]
- Yang C, Wu S, Wu X, et al. Silencing circular RNA UVRAG inhibits bladder cancer growth and metastasis by targeting the microRNA-223/fibroblast growth factor receptor 2 axis. Cancer Sci 2019;110:99-106. [Crossref] [PubMed]
- Izumi H, Kosaka N, Shimizu T, et al. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci 2012;95:4831-41. [Crossref] [PubMed]
- Benmoussa A, Laugier J, Beauparlant CJ, et al. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J Dairy Sci 2020;103:16-29. [Crossref] [PubMed]
- You K, Li SY, Gong J, et al. MicroRNA-125b Promotes Hepatic Stellate Cell Activation and Liver Fibrosis by Activating RhoA Signaling. Mol Ther Nucleic Acids 2018;12:57-66. [Crossref] [PubMed]
- Hyun J, Wang S, Kim J, et al. MicroRNA125b-mediated Hedgehog signaling influences liver regeneration by chorionic plate-derived mesenchymal stem cells. Sci Rep 2015;5:14135. [Crossref] [PubMed]
- Zhou JN, Zeng Q, Wang HY, et al. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology 2015;62:801-15. [Crossref] [PubMed]
- Xu N, Brodin P, Wei T, et al. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol 2011;131:1521-9. [Crossref] [PubMed]
- Qu H, Wu S, Li J, et al. MiR-125b regulates the differentiation of hair follicles in Fine-wool Sheep and Cashmere goats by targeting MXD4 and FGFR2. Anim Biotechnol 2021; [Crossref] [PubMed]
- Csak T, Bala S, Lippai D, et al. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS One 2015;10:e0129251. [Crossref] [PubMed]
- Liu D, Han P, Gao C, et al. microRNA-155 Modulates Hepatic Stellate Cell Proliferation, Apoptosis, and Cell Cycle Progression in Rats With Alcoholic Hepatitis via the MAPK Signaling Pathway Through Targeting SOCS1. Front Pharmacol 2020;11:270. [Crossref] [PubMed]
- Bala S, Csak T, Saha B, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol 2016;64:1378-87. [Crossref] [PubMed]
- Dai W, Zhao J, Tang N, et al. MicroRNA-155 attenuates activation of hepatic stellate cell by simultaneously preventing EMT process and ERK1 signalling pathway. Liver Int 2015;35:1234-43. [Crossref] [PubMed]
- Bi J, Ge S. Potential roles of BMP9 in liver fibrosis. Int J Mol Sci 2014;15:20656-67. [Crossref] [PubMed]
- Liu H, Zhong L, Yuan T, et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int J Mol Med 2018;41:3379-93. [Crossref] [PubMed]
- Wang L, Shi Z, Wang X, et al. Protective effects of bovine milk exosomes against oxidative stress in IEC-6 cells. Eur J Nutr 2021;60:317-27. [Crossref] [PubMed]
- Sun JF, Zhang D, Gao CJ, et al. Exosome-Mediated MiR-155 Transfer Contributes to Hepatocellular Carcinoma Cell Proliferation by Targeting PTEN. Med Sci Monit Basic Res 2019;25:218-28. [Crossref] [PubMed]
- Fu X, Wen H, Jing L, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci 2017;108:620-31. [Crossref] [PubMed]
- Zhang Y, Wei W, Cheng N, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 2012;56:1631-40. [Crossref] [PubMed]
- Chen L, Ming X, Li W, et al. The microRNA-155 mediates hepatitis B virus replication by reinforcing SOCS1 signalling-induced autophagy. Cell Biochem Funct 2020;38:436-42. [Crossref] [PubMed]
- Chen J, Yu Y, Li S, et al. MicroRNA-30a ameliorates hepatic fibrosis by inhibiting Beclin1-mediated autophagy. J Cell Mol Med 2017;21:3679-92. [Crossref] [PubMed]
- targetscan 7.2: GDF2 http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000249598.1&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1
- Song Q, Zhong L, Chen C, et al. miR-21 synergizes with BMP9 in osteogenic differentiation by activating the BMP9/Smad signaling pathway in murine multilineage cells. Int J Mol Med 2015;36:1497-506. [Crossref] [PubMed]
- Torres JL, Novo-Veleiro I, Manzanedo L, et al. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J Gastroenterol 2018;24:4104-18. [Crossref] [PubMed]
- Li X, Chen Y, Wu S, et al. microRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor γ. Mol Med Rep 2015;11:1017-24. [Crossref] [PubMed]
- Yan G, Li B, Xin X, et al. MicroRNA-34a Promotes Hepatic Stellate Cell Activation via Targeting ACSL1. Med Sci Monit 2015;21:3008-15. [Crossref] [PubMed]
- Tian XF, Ji FJ, Zang HL, et al. Activation of the miR-34a/SIRT1/p53 Signaling Pathway Contributes to the Progress of Liver Fibrosis via Inducing Apoptosis in Hepatocytes but Not in HSCs. PLoS One 2016;11:e0158657. [Crossref] [PubMed]
- Feili X, Wu S, Ye W, et al. MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-β1/Smad3 pathway in hepatic stellate cells. Cell Biol Int 2018;42:1370-6. [Crossref] [PubMed]
- Liu Q, Zhang Y, Yang S, et al. PU.1-deficient mice are resistant to thioacetamide-induced hepatic fibrosis: PU.1 finely regulates Sirt1 expression via transcriptional promotion of miR-34a and miR-29c in hepatic stellate cells. Biosci Rep 2017;37:BSR20170926. [Crossref] [PubMed]
- Gao HN, Guo HY, Zhang H, et al. Yak-milk-derived exosomes promote proliferation of intestinal epithelial cells in an hypoxic environment. J Dairy Sci 2019;102:985-96. [Crossref] [PubMed]
- Gao HN, Ren FZ, Wen PC, et al. Yak milk-derived exosomal microRNAs regulate intestinal epithelial cells on proliferation in hypoxic environment. J Dairy Sci 2021;104:1291-303. [Crossref] [PubMed]
- Copple BL, Bai S, Burgoon LD, et al. Hypoxia-inducible factor-1α regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int 2011;31:230-44. [Crossref] [PubMed]
- Zhan L, Huang C, Meng XM, et al. Hypoxia-inducible factor-1alpha in hepatic fibrosis: A promising therapeutic target. Biochimie 2015;108:1-7. [Crossref] [PubMed]
- Wei J, Feng L, Li Z, et al. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother 2013;67:387-92. [Crossref] [PubMed]
- Zhang Z, Zha Y, Hu W, et al. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J Biol Chem 2013;288:37082-93. [Crossref] [PubMed]
- Zhao J, Tang N, Wu K, et al. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS One 2014;9:e108005. [Crossref] [PubMed]
- Huang Q, Zhang X, Bai F, et al. Methyl helicterte ameliorates liver fibrosis by regulating miR-21-mediated ERK and TGF-β1/Smads pathways. Int Immunopharmacol 2019;66:41-51. [Crossref] [PubMed]
- Zhang Z, Gao Z, Hu W, et al. 3,3'-Diindolylmethane ameliorates experimental hepatic fibrosis via inhibiting miR-21 expression. Br J Pharmacol 2013;170:649-60. [Crossref] [PubMed]
- Zhang J, Jiao J, Cermelli S, et al. miR-21 Inhibition Reduces Liver Fibrosis and Prevents Tumor Development by Inducing Apoptosis of CD24+ Progenitor Cells. Cancer Res 2015;75:1859-67. [Crossref] [PubMed]
- Caviglia JM, Yan J, Jang MK, et al. MicroRNA-21 and Dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology 2018;67:2414-29. [Crossref] [PubMed]
- Ning ZW, Luo XY, Wang GZ, et al. MicroRNA-21 Mediates Angiotensin II-Induced Liver Fibrosis by Activating NLRP3 Inflammasome/IL-1β Axis via Targeting Smad7 and Spry1. Antioxid Redox Signal 2017;27:1-20. [Crossref] [PubMed]
- Li L, Jiang D. Hypoxia-responsive miRNA-21-5p inhibits Runx2 suppression by targeting SMAD7 in MC3T3-E1 cells. J Cell Biochem 2019;120:16867-75. [Crossref] [PubMed]
- Arumugam B, Balagangadharan K, Selvamurugan N. Syringic acid, a phenolic acid, promotes osteoblast differentiation by stimulation of Runx2 expression and targeting of Smad7 by miR-21 in mouse mesenchymal stem cells. J Cell Commun Signal 2018;12:561-73. [Crossref] [PubMed]
- Makhmudi A, Kalim AS. Gunadi. microRNA-21 expressions impact on liver fibrosis in biliary atresia patients. BMC Res Notes 2019;12:189. [Crossref] [PubMed]
- Shen W, Chen G, Dong R, et al. MicroRNA-21/PTEN/Akt axis in the fibrogenesis of biliary atresia. J Pediatr Surg 2014;49:1738-41. [Crossref] [PubMed]
- Zhou Y, Ren H, Dai B, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res 2018;37:324. [Crossref] [PubMed]
- Wu N, McDaniel K, Zhou T, et al. Knockout of microRNA-21 attenuates alcoholic hepatitis through the VHL/NF-κB signaling pathway in hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol 2018;315:G385-98. [Crossref] [PubMed]
- Sun J, Shi L, Xiao T, et al. microRNA-21, via the HIF-1α/VEGF signaling pathway, is involved in arsenite-induced hepatic fibrosis through aberrant cross-talk of hepatocytes and hepatic stellate cells. Chemosphere 2021;266:129177. [Crossref] [PubMed]
- He G, Ding J, Zhang Y, et al. microRNA-21: a key modulator in oncogenic viral infections. RNA Biol 2021;18:809-17. [Crossref] [PubMed]
- Baddela VS, Nayan V, Rani P, et al. Physicochemical Biomolecular Insights into Buffalo Milk-Derived Nanovesicles. Appl Biochem Biotechnol 2016;178:544-57. [Crossref] [PubMed]
- Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014;146:1513-24. [Crossref] [PubMed]
- Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol 2020;72:558-77. [Crossref] [PubMed]
- Tsiaoussis GI, Assimakopoulos SF, Tsamandas AC, et al. Intestinal barrier dysfunction in cirrhosis: Current concepts in pathophysiology and clinical implications. World J Hepatol 2015;7:2058-68. [Crossref] [PubMed]
- Wang L, Llorente C, Hartmann P, et al. Methods to determine intestinal permeability and bacterial translocation during liver disease. J Immunol Methods 2015;421:44-53. [Crossref] [PubMed]
- Pijls KE, Jonkers DM, Elamin EE, et al. Intestinal epithelial barrier function in liver cirrhosis: an extensive review of the literature. Liver Int 2013;33:1457-69. [Crossref] [PubMed]
- Pijls KE, Koek GH, Elamin EE, et al. Large intestine permeability is increased in patients with compensated liver cirrhosis. Am J Physiol Gastrointest Liver Physiol 2014;306:G147-53. [Crossref] [PubMed]
- Assimakopoulos SF, Tsamandas AC, Tsiaoussis GI, et al. Altered intestinal tight junctions' expression in patients with liver cirrhosis: a pathogenetic mechanism of intestinal hyperpermeability. Eur J Clin Invest 2012;42:439-46. [Crossref] [PubMed]
- Fukui H. Role of Gut Dysbiosis in Liver Diseases: What Have We Learned So Far? Diseases 2019;7:58. [Crossref] [PubMed]
- Tulkens J, Vergauwen G, Van Deun J, et al. Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 2020;69:191-3. [Crossref] [PubMed]
- Tranah TH, Edwards LA, Schnabl B, et al. Targeting the gut-liver-immune axis to treat cirrhosis. Gut 2021;70:982-94. [Crossref] [PubMed]
- Hock A, Miyake H, Li B, et al. Breast milk-derived exosomes promote intestinal epithelial cell growth. J Pediatr Surg 2017;52:755-9. [Crossref] [PubMed]
- Martin C, Patel M, Williams S, et al. Human breast milk-derived exosomes attenuate cell death in intestinal epithelial cells. Innate Immun 2018;24:278-84. [Crossref] [PubMed]
- Miyake H, Lee C, Chusilp S, et al. Human breast milk exosomes attenuate intestinal damage. Pediatr Surg Int 2020;36:155-63. [Crossref] [PubMed]
- Pisano C, Galley J, Elbahrawy M, et al. Human Breast Milk-Derived Extracellular Vesicles in the Protection Against Experimental Necrotizing Enterocolitis. J Pediatr Surg 2020;55:54-8. [Crossref] [PubMed]
- Xie MY, Chen T, Xi QY, et al. Porcine milk exosome miRNAs protect intestinal epithelial cells against deoxynivalenol-induced damage. Biochem Pharmacol 2020;175:113898. [Crossref] [PubMed]
- Dong P, Zhang Y, Yan DY, et al. Protective Effects of Human Milk-Derived Exosomes on Intestinal Stem Cells Damaged by Oxidative Stress. Cell Transplant 2020;29:963689720912690. [Crossref] [PubMed]
- Tong L, Hao H, Zhang X, et al. Oral Administration of Bovine Milk-Derived Extracellular Vesicles Alters the Gut Microbiota and Enhances Intestinal Immunity in Mice. Mol Nutr Food Res 2020;64:e1901251. [Crossref] [PubMed]
- He S, Liu G, Zhu X. Human breast milk-derived exosomes may help maintain intestinal epithelial barrier integrity. Pediatr Res 2021;90:366-72. [Crossref] [PubMed]
- Zeng R, Wang J, Zhuo Z, et al. Stem cells and exosomes: promising candidates for necrotizing enterocolitis therapy. Stem Cell Res Ther 2021;12:323. [Crossref] [PubMed]
- Maghraby MK, Li B, Chi L, et al. Extracellular vesicles isolated from milk can improve gut barrier dysfunction induced by malnutrition. Sci Rep 2021;11:7635. [Crossref] [PubMed]
- Yu S, Zhao Z, Xu X, et al. Characterization of three different types of extracellular vesicles and their impact on bacterial growth. Food Chem 2019;272:372-8. [Crossref] [PubMed]
- Zhou F, Paz HA, Sadri M, et al. Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am J Physiol Gastrointest Liver Physiol 2019;317:G618-24. [Crossref] [PubMed]
- Fan K, Wu K, Lin L, et al. Metformin mitigates carbon tetrachloride-induced TGF-β1/Smad3 signaling and liver fibrosis in mice. Biomed Pharmacother 2017;90:421-6. [Crossref] [PubMed]
- Feng YH, Wu CL, Shiau AL, et al. MicroRNA-21-mediated regulation of Sprouty2 protein expression enhances the cytotoxic effect of 5-fluorouracil and metformin in colon cancer cells. Int J Mol Med 2012;29:920-6. [PubMed]
- Kalogirou C, Schäfer D, Krebs M, et al. Metformin-Derived Growth Inhibition in Renal Cell Carcinoma Depends on miR-21-Mediated PTEN Expression. Urol Int 2016;96:106-15. [Crossref] [PubMed]
- Sun R, Ma X, Cai X, et al. The effect and mechanism of action of metformin on in vitro FaDu cell proliferation. J Int Med Res 2016;44:1049-54. [Crossref] [PubMed]
- Pulito C, Mori F, Sacconi A, et al. Metformin-induced ablation of microRNA 21-5p releases Sestrin-1 and CAB39L antitumoral activities. Cell Discov 2017;3:17022. [Crossref] [PubMed]
- Deng Y, Ma W. Metformin inhibits HaCaT cell viability via the miR-21/PTEN/Akt signaling pathway. Mol Med Rep 2018;17:4062-6. [PubMed]
- Melnik BC, John SM, Carrera-Bastos P, et al. MicroRNA-21-Enriched Exosomes as Epigenetic Regulators in Melanomagenesis and Melanoma Progression: The Impact of Western Lifestyle Factors. Cancers (Basel) 2020;12:2111. [Crossref] [PubMed]
- Khokhar M, Roy D, Bajpai NK, et al. Metformin mediates MicroRNA-21 regulated circulating matrix metalloproteinase-9 in diabetic nephropathy: an in-silico and clinical study. Arch Physiol Biochem 2021;1-11. [Crossref] [PubMed]
- Alimoradi N, Firouzabadi N, Fatehi R. How metformin affects various malignancies by means of microRNAs: a brief review. Cancer Cell Int 2021;21:207. [Crossref] [PubMed]
- Belloni L, Di Cocco S, Guerrieri F, et al. Targeting a phospho-STAT3-miRNAs pathway improves vesicular hepatic steatosis in an in vitro and in vivo model. Sci Rep 2018;8:13638. [Crossref] [PubMed]
- Shu C, Yan D, Chen C, et al. Metformin exhibits its therapeutic effect in the treatment of pre-eclampsia via modulating the Met/H19/miR-148a-5p/P28 and Met/H19/miR-216-3p/EBI3 signaling pathways. Int Immunopharmacol 2019;74:105693. [Crossref] [PubMed]
- Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013;52:101-12. [Crossref] [PubMed]
- Yan L, Zhou J, Gao Y, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 2015;34:3076-84. [Crossref] [PubMed]
- Zhong T, Men Y, Lu L, et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene 2017;36:2345-54. [Crossref] [PubMed]
- Chen Z, Wei H, Zhao X, et al. Metformin treatment alleviates polycystic ovary syndrome by decreasing the expression of MMP-2 and MMP-9 via H19/miR-29b-3p and AKT/mTOR/autophagy signaling pathways. J Cell Physiol 2019;234:19964-76. [Crossref] [PubMed]
- Zhu J, Luo Z, Pan Y, et al. H19/miR-148a/USP4 axis facilitates liver fibrosis by enhancing TGF-β signaling in both hepatic stellate cells and hepatocytes. J Cell Physiol 2019;234:9698-710. [Crossref] [PubMed]
- Heo MJ, Kim YM, Koo JH, et al. microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget 2014;5:2792-806. [Crossref] [PubMed]
- Zhang L, Xing M, Wang X, et al. MiR-148a suppresses invasion and induces apoptosis of breast cancer cells by regulating USP4 and BIM expression. Int J Clin Exp Pathol 2017;10:8361-8. [PubMed]
- McCarty MF. Metformin may antagonize Lin28 and/or Lin28B activity, thereby boosting let-7 levels and antagonizing cancer progression. Med Hypotheses 2012;78:262-9. [Crossref] [PubMed]
- Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci U S A 2013;110:972-7. [Crossref] [PubMed]
- Balzeau J, Menezes MR, Cao S, et al. The LIN28/let-7 Pathway in Cancer. Front Genet 2017;8:31. [Crossref] [PubMed]
- Tao L, Fan X, Sun J, et al. Metformin prevented high glucose-induced endothelial reactive oxygen species via OGG1 in an AMPKα-Lin-28 dependent pathway. Life Sci 2021;268:119015. [Crossref] [PubMed]
- Melnik BC, Schmitz G. Metformin: An inhibitor of mTORC1 signaling. J Endocrinol Diabetes Obes 2014;2:1029.
- Sato Y, Qiu J, Hirose T, et al. Metformin slows liver cyst formation and fibrosis in experimental model of polycystic liver disease. Am J Physiol Gastrointest Liver Physiol 2021;320:G464-73. [Crossref] [PubMed]
- Li J, Hernanda PY, Bramer WM, et al. Anti-tumor effects of metformin in animal models of hepatocellular carcinoma: a systematic review and meta-analysis. PLoS One 2015;10:e0127967. [Crossref] [PubMed]
- Cunha V, Cotrim HP, Rocha R, et al. Metformin in the prevention of hepatocellular carcinoma in diabetic patients: A systematic review. Ann Hepatol 2020;19:232-7. [Crossref] [PubMed]
- Dhar D, Baglieri J, Kisseleva T, et al. Mechanisms of liver fibrosis and its role in liver cancer. Exp Biol Med (Maywood) 2020;245:96-108. [Crossref] [PubMed]
- Melnik BC. Dairy consumption and hepatocellular carcinoma risk. Ann Transl Med 2021;9:736. [Crossref] [PubMed]
- Gay MD, Safronenka A, Cao H, et al. Targeting the Cholecystokinin Receptor: A Novel Approach for Treatment and Prevention of Hepatocellular Cancer. Cancer Prev Res (Phila) 2021;14:17-30. [Crossref] [PubMed]
- Zhao Z, Lin CY, Cheng K. siRNA- and miRNA-based therapeutics for liver fibrosis. Transl Res 2019;214:17-29. [Crossref] [PubMed]
- Aqil F, Munagala R, Jeyabalan J, et al. Milk exosomes - Natural nanoparticles for siRNA delivery. Cancer Lett 2019;449:186-95. [Crossref] [PubMed]
- Myrzabekova M, Labeit S, Niyazova R, et al. Identification of bovine miRNAs with the potentital to affect human gene expression. Front Genet 2022;12:705350. [Crossref] [PubMed]
- Aarts J, Boleij A, Pieters BCH, et al. Flood Control: How Milk-Derived Extracellular Vesicles Can Help to Improve the Intestinal Barrier Function and Break the Gut-Joint Axis in Rheumatoid Arthritis. Front Immunol 2021;12:703277. [Crossref] [PubMed]
- Zhong J, Xia B, Shan S, et al. High-quality milk exosomes as oral drug delivery system. Biomaterials 2021;277:121126. [Crossref] [PubMed]
- Börger V, Bremer M, Ferrer-Tur R, et al. Mesenchymal Stem/Stromal Cell-Derived Extracellular Vesicles and Their Potential as Novel Immunomodulatory Therapeutic Agents. Int J Mol Sci 2017;18:1450. [Crossref] [PubMed]
- Zhang C, Lu X, Hu J, et al. Bovine Milk Exosomes Alleviate Cardiac Fibrosis via Enhancing Angiogenesis In Vivo and In Vitro. J Cardiovasc Transl Res 2021; [Crossref] [PubMed]
- Wang H, Wang B, Zhang A, et al. Exosome-Mediated miR-29 Transfer Reduces Muscle Atrophy and Kidney Fibrosis in Mice. Mol Ther 2019;27:571-83. [Crossref] [PubMed]
Cite this article as: Melnik BC, Weiskirchen R, Stremmel W, John SM, Schmitz G. Narrative review of bovine milk exosomes as a potential therapeutic option for hepatic fibrosis. Dig Med Res 2022;5:14.


