
Paediatric IBD: the host, diet & microbes in pathogenesis & treatment: a narrative review
Introduction: paediatric inflammatory bowel diseases
The global prevalence of inflammatory bowel diseases (IBD) has been steadily increasing and is now greater than 0.3% (1). In some Western countries, such as Canada, rates are projected to be as high as 1% by the year 2030 (2). The prevalence in children has also been increasing (3), with up to 25% of diagnoses occurring in the paediatric population (4,5). There are two main subtypes of IBD, including Crohn disease (CD) and ulcerative colitis (UC). IBD consists of chronic intestinal inflammation, which presents with skip lesions occurring anywhere along the gastrointestinal tract from mouth to anus in CD, while in UC inflammation is restricted to the colon in a contiguous fashion. The aetiology of IBD is not completely understood, and there is currently no cure; however, there is evidence that genetics, environment, immune response, and the intestinal microbiome interact to play a role in disease development and progression (6-14). Growth and development concerns are unique to paediatric IBD (pIBD), including failure to thrive, which is a common presentation that leads to an IBD diagnosis; furthermore, pIBD carries a risk of linear growth failure, delayed puberty, and reduced peak bone density (15-17). Children with IBD are also at an increased risk of psychosocial difficulties including depression and anxiety (18).
Younger age of diagnosis in paediatric CD (pCD) is associated with more complicated disease (19,20). Complicated disease is defined as stricturing and/or penetrating disease in pCD, and is associated with poorer outcomes and increased need for surgery (21). pCD patients are also more likely to have small bowel involvement than adults (42% compared to 15% of adults) (22). Paediatric UC (pUC) patients are more likely to have extensive inflammation in the form of pancolitis, ~43–81% (22-24) compared to ~20–35% in adults (22,25,26). Almost half of children diagnosed with pCD, and roughly 16% of children diagnosed with pUC require surgery in the ten years following diagnosis (27), and children with IBD have on average a lower health-related quality of life (28). In follow-up studies, diagnosis with IBD in childhood has been associated with increased risk of cancer and mortality in adulthood (29). To improve outcomes and prevent the complications of disease associated with a young age at diagnosis, there is a need to better define causes and predictors of response to therapy in pIBD. We present the following article in accordance with the narrative review checklist (available at http://dx.doi.org/10.21037/dmr-20-160).
Methods
The purpose of this literature review is to elucidate the host-microbe interactions in pIBD and describe how these interactions are influenced by diet and current disease therapies. Original, peer-reviewed, English language research published in the most recent five years was considered first and foremost, with the addition of older studies if they provided novel, foundational, or relevant information. Human studies were prioritized, followed by animal and then in vitro evidence, respectively.
Aetiology & pathophysiology: host, microbe, and diet
The pathogenesis of pIBD is complex, involving genetic and environmental factors. To clearly define how microbes and diet are involved in pathogenesis, and how they could therefore contribute to therapy response, we will first provide an overview of current knowledge of disease pathogenesis in pIBD in general. This will include details on genetics, microbes, the intestinal barrier, metabolites, and diet.
IBD-associated genes
Although genetics plays a role in pIBD, most patients are diagnosed without a family history (30), suggesting that genetic factors are less significant than environmental ones in the pathogenesis. Graham and Xavier provide a detailed review of IBD genetics (31). Associated genes are involved in intestinal epithelial barrier integrity, microbial recognition and clearance, and regulation of the host-immune response (32,33). The caspase recruitment domain family member 15 gene (CARD15), known as nucleotide-binding oligomerization domain two (NOD2), displays the strongest genetic association with IBD, and is specifically correlated with CD (34). NOD2 is a pattern recognition molecule in the cell cytosol that can recognize bacterial peptidoglycan (35,36). Individuals with the IBD-associated NOD2 variants have decreased intestinal antimicrobial peptides (e.g., α-defensins) (37), and the most common IBD-associated variant has no response in vitro to bacterial peptidoglycan and lipopolysaccharide (LPS) (38), along with a drastically reduced ability to prevent intracellular invasion (39). Patients with these variants also have lower circulating levels of anti-tumour necrosis factor-α (TNF-α) monoclonal antibody (MAb) (discussed below) between doses than patients with the wild-type gene, which may result in reduced treatment efficacy (40). Children with NOD2 variants are also more likely to present with growth impairment (41). Another IBD-associated variant is in the CARD9 gene, which results in impaired innate immune response to intracellular pathogens and increased pro-inflammatory signalling (including TNF-α) through increased nuclear factor-κB (NF-κB) signalling (42,43). Individuals with these variants also exhibit increased Malassezia restricta, a fungus capable of exacerbating colitis in animal models (44). Inability to adequately clear microbes, as may occur with the IBD-associated NOD2 and CARD9 variants, could result in chronic inflammation as microbes persist inappropriately in the intracellular or juxta-epithelial intestinal environment.
There are also IBD-associated genetic variants linked to increased intestinal permeability in IBD, such as Janus kinase (JAK)2 in CD (45). JAK2 is involved in cytokine signalling (46) and its inhibition can decrease T cell-mediated inflammation (47). The JAK inhibitor Tofacitinib has proven effective in some IBD populations (48), potentially by reducing JAK-driven intestinal permeability, reducing antigen translocation from the intraluminal environment, and production of T cell-mediated inflammation. Other examples of genetic associations with reduced barrier integrity include Prostaglandin E Receptor 4 (PTGER4) in CD and Hepatocyte Nuclear 4 Alpha (HNF4A) in UC (49), regulating junctional/cytoskeletal proteins and claudin-15 (important for epithelial tight junctions), respectively (50). Additional IBD-associated variants in genes involved in cell adhesion and tight junctions include C1orf106 and RNF186 (51-53). Suboptimal intestinal barrier function may result in increased antigen exposure and perpetuate a pro-inflammatory immune response (54-56).
Although a significant number of genetic associations have been identified in IBD, to date only 26% and 19% of the heritability of CD and UC, respectively, can be explained by genetics (57). Genetic associations also cannot explain the recent rises in incidence (2), highlighting the importance of assessing environmental factors as contributors to the pathophysiology of IBD.
Microbial involvement in IBD
The human gut microbiome is composed of bacteria, viruses, archaea, and eukaryotes, with approximately as many bacterial cells as there are human cells in the body (58). Alterations in the gut microbiome are thought to play important roles in numerous human diseases. The microbiome is also critical to health, and is essential for healthy immune development and certain metabolic functions, such as dietary fibre and protein fermentation, and generation of certain vitamins and neurotransmitters (59-61). The normal intestinal bacterial microbiota is composed mostly of obligate anaerobes from several main phyla, including: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and trace Verrucomicrobia, Fusobacteria, Cyanobacteria, and candidate division TM7 (62). The role of viruses, fungi, and other eukaryotes in the intestinal microbiome has been relatively understudied, but evidence suggests a dysbiosis of fungi and viruses also exists in paediatric (63-65) and adult (66,67) IBD. Interest in the role of fungi, which constitute a relatively small proportion of the total intestinal microbiota (68), was sparked by the increased prevalence of anti-fungal antibodies in patients with IBD relative to controls (69). The IBD mycobiome is characterized by reduced diversity and an increased total fungal load, with increased ratios of Basidiomycota to Ascomycota, and a decrease in the ratio of Saccharomyces cerevisiae to Candida albicans (44,63,67). There is also an increase in the previously mentioned M. restrica, seen with IBD-associated CARD9 variants (44,70). Alterations in fungi total and relative abundances and diversity, and potential related therapeutics were recently reviewed by Lam et al. (70). Changes in the intestinal virome in IBD are even less understood (71,72), but have been shown to trigger colitis in some animal IBD models (73), and likely influence complex relationships within the microbial ecosystem (74), including the effects of bacteriophage on the bacterial microbiota (75,76). Potential implications of the intestinal virome are discussed in several specific reviews (77,78).
Alterations in the intestinal bacterial microbiota are a well-documented hallmark of IBD, with somewhat different alterations in CD vs. UC, and more significant differences with more active disease, relative to controls (79). The microbial dysbiosis in IBD is likely, in-part, independent of associated host genetic factors, as demonstrated by numerous twin studies (80,81). Microbiota differences between twins concordant for CD supports a likely role for the influence by environmental factors in establishing and maintaining dysbiosis (82,83). Environmental factors known to play a role in influencing the intestinal microbiota include antibiotic exposure (84,85), infections (including COVID-19) (86), nutrition, and other environmental exposures such as tobacco smoke (87-93). Microbial changes in IBD show decreases in strict anaerobic species within the Firmicutes and Actinobacteria phyla, with increases in oxygen tolerant Proteobacteria and Bacteroidetes sp. (94). Other changes include lower levels of mucosal bacteria species richness in pCD that is not always reflected in stool analyses (95), reduced faecal microbiota diversity (96), and alterations in certain mucosal and bacterial taxa, including higher numbers of Enterobacteriaceae and lower abundances of strictly anaerobic Firmicutes, such as Clostridiales (95). Reduced levels of Clostridium sp., Faecalibacterium prausnitzii, and Bifidobacterium sp. are seen in IBD, with relative abundances reduced more in CD than UC (79). Reduced Bacteroides species are also seen in IBD, more so in active disease than remission (97). F. prausnitzii, an important fibre fermenter, has been shown to have anti-inflammatory effects, and its absence is predictive of disease relapse (98). Beneficial microbes such as F. prausnitzii that are reduced in IBD may improve mucosal barrier function through tight junction proteins, inducible heat shock proteins, immunomodulation, and even production of anti-inflammatory peptides (99-103).
Microbial dysbiosis correlates strongly with disease severity, as measured by the paediatric Crohn disease activity index (PCDAI) (95). Microbes may even predict future disease activity: levels of Enterobacteriaceae, Fusobacterium, and Haemophilus, when combined with age of disease onset and PCDAI at diagnosis, have been found to be significantly predictive of future PCDAI (95). Unaffected siblings of CD patients also display an altered intestinal microbiome compared to controls that correlates with their increased risk for developing IBD, suggesting a causative role for the microbiome in IBD pathogenesis (104-106). Disease location is also associated with variations in microbial abundance: patients with predominantly ileal CD had lower abundances of F. prausnitzii and higher levels of Escherichia coli in intestinal mucosa specimens compared to patients with predominantly colonic/ileocolonic CD (81).
Although UC has shown less microbial distinction from non-IBD controls than CD (107), Shah et al. found that patient response to initial therapy was associated with differential abundances of certain microbial taxa (108). Certain microbes may predict or even mediate a patient’s future response to treatment in pUC, as Shah et al. found that certain Clostridium and Bacteroides OTUs were only detectable at baseline in the pUC patients who went on to have no clinical response to treatment (108). Similarly, high Candida abundance at baseline was associated with clinical response and increased bacterial diversity following faecal microbiota transplantation in active UC (109). In severe pUC, phylum level changes included lower levels of Firmicutes and higher levels of Proteobacteria compared to controls. The lower microbial richness in pUC was more prominent in children who failed to respond to glucocorticoid therapy (110). Additionally, fluctuations in the UC microbiome are associated with disease severity and need for colectomy (111), and significant depletion of Ruminococcaeae and Lachnospiraceae with an increased abundance of Streptococcus anginosus were found in patients with more severe disease.
While altered microbiota correlating with disease severity is well documented at various taxonomic levels (94), there is yet to be any specific microbial culprit linked with disease flares. Sequencing comparing mucosal microbiota in inflamed vs. non-inflamed bowel segments has not identified specific taxa associated with inflammation across individuals but does show significant composition changes in the non-inflamed terminal ileum of pUC patients, suggesting microbial alterations upstream of local inflammation (112,113). Likely, complex host-microbe interactions are specific to each individual. Roy et al. have shown that specific variations in microbial dysbiosis introduced in animal models lead to unique colitis phenotypes and pathologies specific to the microbial community introduced (114). Future techniques to identify ‘problematic microbiota’ unique to each patient with IBD may involve identifying levels of immunoglobulin (Ig) binding found on microbes isolated from patients. IgA and IgG have shown differential microbial binding in patients with IBD, including differential binding between UC and CD (115,116). High levels of Ig coating has been used to identify bacteria from IBD patients that induce inflammation in animal and in vitro models, suggesting that these techniques could be used to identify culprit bacteria in specific patients (117,118). Mucosal IgA can help reduce microbial invasion (119). In some animal models certain antibodies against the intestinal microbiota protect against bacterial sepsis (120); the role of these antibodies in IBD remains unclear but has been explored as a marker of inappropriate immune responses to gut bacteria (121) or increased antigen exposure through intestinal barrier disruption with increased disease activity (115,122,123). Some antibodies directed against commensal microbes and fungi, such as anti-Saccharomyces cerevisiae (ASCA), have limited diagnostic and predictive roles in IBD (124).
Altered intestinal barrier in IBD
Impaired barrier integrity is another feature of IBD (125-129). pUC patients show reduced mucous barrier thickness, and fewer mucin-containing goblet cells, with bacteria colonizing closer to intestinal epithelial cells than controls (113). Similarly, bacterial colonization was in closer proximity to intestinal epithelial cells in pCD vs. controls (113). Confocal laser endomicroscopy has shown increased leakage of IV-administered fluorescein through the intestinal epithelial barrier in pCD and pUC compared to controls (125); in adults, increased fluorescein leakage was associated with disease relapse (55). As mentioned in relation to patient genetics, an impaired epithelial barrier may result in increased bacterial and antigen translocation that perpetuates chronic inflammation.
Alterations in the intestinal microbiota and host barrier integrity seen in IBD carry a number of potential consequences for the patient (130). Microbes can directly impact health by increasing or decreasing resistance to colonization by pathogens, producing vitamins and nutrients, and training the development of a balanced immune system (130-132). A balanced host-microbe interaction is essential for health; germ-free mice show aberrations in a number of body systems including: cardiovascular, respiratory, gastrointestinal, and immunity (131). Germ-free animals also show significant morphological and functional intestinal changes (131). Some of these aberrances can be corrected by colonization with specific bacteria, including improving intestinal barrier integrity with the addition of certain Lactobacillus sp. to germ-free animals (133-135). An unhealthy host-microbe relationship in IBD may perpetuate or trigger disease, and elucidation of this relationship can improve our understanding of the aetiologies of these diseases.
Metabolites: linking diet to microbes in IBD
Metabolites are compounds necessary for or formed during metabolism and represent dietary, host, and microbe factors reflecting the real-time microenvironment of the gut. Identification and quantification of metabolites can provide a concrete assessment of microbial function and human metabolism. In addition to altered microbial abundances, variations in urinary metabolites suggest altered microbial function in the IBD microbiome. In adults with IBD, serum and urine metabolites can distinguish active disease from remission (10). Urine metabolites can discriminate adult IBD patients from healthy controls, with a varying ability to distinguish CD from UC (136,137), while serum and faecal metabolites are capable of distinguishing pCD from pUC and healthy controls (138). A limited number of studies suggest that urinary metabolomics can also differentiate pIBD from healthy controls (139). Metabolomics of mucosal surface samples collected during colonoscopy in pIBD patients linked luminal succinate to bacterial invasion in vitro, highlighting the importance of the gut microenvironment in IBD (140). Lavelle & Sokol detail the groups of microbe-associated metabolites significant in IBD and their potential roles (141).
Metabolites commonly altered in paediatric and adult IBD belong to metabolic pathways suggesting variations in glutathione metabolism, the citric acid cycle (CAC), and importantly, microbial activity (136,139,142). Hippurate is defined as a co-metabolite, as it is produced by a combination of host and microbial metabolism (143); it is notably absent in germ-free animals (143), and significantly reduced in IBD (144). Hippurate has been positively associated with microbiome diversity, as well as increased fruit and whole grain intake (145). When potential dietary influences were excluded, hippurate persisted as a marker for IBD vs. controls (146). Supplementation with the hippurate precursor sodium benzoate does not increase urinary hippurate in CD or controls, and CD patients have persistently lower levels, suggesting an absence of the microbial metabolic function (147).
Other microbe-associated metabolites include short chain fatty acids (SCFA), which are produced from microbial fermentation of dietary fibres, and to a lesser extent amino acids; these are often significantly reduced in both adult and paediatric IBD (136,137,148,149). Stool from IBD patients typically has less acetate, butyrate, and propionate, while lactate and pyruvate are increased (148). Effects of SCFA include suppressing intestinal permeability and increasing the population of regulatory T-cells that can help to prevent chronic inflammation (150,151). SCFA can also activate anti-inflammatory signalling cascades by binding to intestinal G-protein coupled receptors such as GPR109A, GPR43, and GPR41 (152). Impaired metabolism of SCFA has been implicated in IBD; for example, experimental dextran sulphate sodium (DSS)-induced colitis is characterized by reduced butyrate oxidation and increased glucose oxidation (153). Additionally, in one study, GPR43 expression in the ileum was significantly reduced in CD compared to controls, regardless of disease activity (154). Inflammation impacts the response to butyrate in IBD; ex vivo organoids from IBD patients and controls respond similarly to butyrate, but when the pro-inflammatory cytokine TNF-α is added butyrate uptake is reduced in IBD organoids only (155). This could explain why exogenous administration of SCFAs has not consistently resulted in clinical improvement in the treatment of IBD (156). Previous studies examining direct administration of SCFA (using butyrate enemas) have shown mixed results, despite other promising in vitro, ex vivo, and animal studies showing decreased inflammatory markers and improved intestinal barrier (157,158).
Butyrate produced by bacterial fermentation is the primary fuel source for colonocytes (159); not surprisingly, colonocytes from germ-free mice have fewer CAC intermediate enzymes (153). Butyrate has multiple physiologic effects, including reducing bacterial translocation in vitro (99), and can act as both an energy source and a histone deacetylase (HDAC) inhibitor (153). Butyrate concentrations are increased by some resistant starches (RS) (160), and SCFA production varies in response to diet and intestinal transit time (160). Stool SCFA content does not always reflect production through fermentation, as longer transit time is associated with reduced butyrate (160). Impacts of transit time on SCFA concentrations could be due to increased opportunity for absorption and utilization during slower transit, or related to delivery time of RS and dietary fibre to the large bowel and time for microbial metabolism and fermentation (160). A number of other factors are known to impact SCFA concentrations; antibiotics reduce SCFA and intake of different dietary fibres can increase SCFAs in varying amounts (161). Additionally, SCFA production can modify the intestinal microbiome: their acidity can select for more acid-tolerant bacteria (161). SCFA may play a role in patient response to treatment; Wang et al. found a significant reduction in SCFA-producing bacteria prior to Infliximab (IFX) therapy in pIBD, with a shift to a SCFA profile more similar to healthy controls after therapy, and that the abundance of SCFA-producing species was predictive of sustained remission in pIBD (158). However, it is important to recognize the limitations of metabolomics, especially as this relates to interindividual variation and lack of standardized protocols.
Environment: focus on diet
A “Western” diet that is low in dietary fibre and high in processed and fatty foods has been proposed to partially account for the recent rise in the incidence in IBD, particularly in the western world (162). Although a number of dietary risk factors and associations have been recognised, the majority have been identified through case-control or cohort studies. It is difficult to design and conduct experimental dietary studies that can prove causality, and in animal studies it can be challenging to approximate a human diet and consumption patterns. Assessing for associations with individual foods or nutrients may miss capturing the complexity of micro and macronutrient interactions. Additional complexities include the associations of dietary intake with culture and genetics, as well as numerous reporting biases.
Diet is associated with both increased and decreased risk in adult and paediatric IBD. CD has especially strong associations with dietary intake (163). Foods associated with increased risk of IBD have included fast-food (164) and increased protein intake that is potentially specific to animal protein (163,165-167). Protective factors include breast-feeding (163), and specifically for pCD: increased vegetables, fruits, fish, olive oil, grains, and nuts (168). Some sex-specific associations have been identified, and foods associated with increased risk for pCD in females included meat, sugary foods, and high fat foods (168). Many of these studies were conducted prospectively, for example E3N (166), EPIC (169), the Nurses’ Health Study I & II, and Health Professionals Follow-up Study (170), suggesting that dietary intakes may establish an intestinal environment that favours development of IBD. Diet is proposed to exert effects through alterations in host immunity, host barrier integrity, and alterations in the microbiota, including viruses (163,171). Potential mechanisms are discussed in depth in the review by Levine et al.; we will discuss them briefly here (163). Host variability in these factors, specifically the microbiota, likely plays a significant role in the complex associations seen with certain dietary components, such as meat or dietary fibres. In the absence of dietary fibres, some microbes display enhanced mucolytic activity – digesting and depleting the host intestinal mucous layer, compromising intestinal barrier integrity, and resulting in increased immune activation (163). Resulting epithelial barrier disruption, as evidenced by increased gap density, correlates positively with increased inflammation and disease activity (125). Dietary fibres and RS can increase SCFA production, potentially decreasing intestinal permeability (163). Other diet components may also impact the levels of SCFA receptors; in mice fed a high fat and sugar diet there was reduced GPR43 receptor expression (154).
Diet is one of the main determinants of the intestinal microbiome (172). Previous studies have found that dietary patterns are significantly more predictive of the intestinal microbiome than individual nutrients—likely as the complex interaction of whole foods cannot be predicted from isolated components (93). In mice, a Western style diet high in fat and sugar increased colonization with pathogenic organisms such as adherent invasive E. coli (AIEC) (173), which is a key pathobiont implicated in the pathophysiology of CD (174). Targeting these associations between host, diet, and microbe, a number of dietary therapies have been developed, and are described in the Treatment section.
Diet-host-microbe interactions in IBD
The genetic, microbial, dietary, and metabolic associations in IBD suggest that dysregulated host-microbe relationships, further influenced by diet, are important in the pathogenesis of IBD. This is additionally supported by evidence that microbiota-targeting therapies, like antibiotics and diet, can result in clinical improvement in IBD (99,163). Faecal microbiota transplant (FMT) is an emerging microbe-based therapy that has shown some promise, but experience in children is limited (175). A variety of protocols exist but evidence is conflicting on many aspects, such as: donor selection (universal may be beneficial) (176,177), fresh vs. previously frozen donor stool (178), or need for antibiotic pre-treatment (178-180). Baseline patient microbiota is likely a factor in the variability of outcomes with FMT; in adult UC, there is evidence that the baseline mycobiome, specifically increased Candida abundance, can be predictive of a positive FMT response (181). Much of the evidence for FMT is specific to treatment of a superimposed Clostridioides difficile infection (CDI) (182). FMT for CDI in IBD is less effective than without IBD and results in fewer microbial shifts (183,184). Although most adverse events with FMT in IBD are mild and include non-specific symptoms such as diarrhoea, nausea, and abdominal pain (176), some potentially serious disease flares may occur (182,185). FMT in IBD is further discussed in several recent reviews (179,186-189).
As another example, diversion of the faecal stream in pIBD through an ileostomy can result in clinical improvement, with disease severity often increasing again when the faecal stream is returned to the colon (190). Additionally, almost all colitis animal models, such as interleukin-10 (IL-10) knockout or TNFΔARE mice, do not develop intestinal inflammation under germ-free conditions (191,192).
Increased leakage of luminal antigens across the intestinal barrier in IBD, presumably due to genetic and environmental/diet factors, results in an excessive cytokine-dependent inflammatory response in mucosal tissue (193). These antigens can include bacterial products or components such as LPS or peptidoglycan. Microbial antigens can increase NF-κB signalling through increased production of TNF-α, which is recognized as an integral part of protection against invading pathogens (194). TNF-α is thought to play a role in the pathogenesis of IBD via fibroblast activation, enhanced production of pro-inflammatory cytokines, and enhanced T-cell resistance to apoptosis (193). TNF-α effects through NF-κB also result in Paneth cell death, damage to intestinal epithelial cells, and increased production of matrix metalloproteinases (MMPs) by myofibroblasts (193). Paneth cells are specialized secretory intestinal epithelial cells located in intestinal crypts of the small bowel (but also can be found in the large bowel in IBD) that are important for producing antimicrobial and immune-stimulating molecules (195). They have been shown to be important in defence against intestinal pathogens and play a role in regulating the abundance of certain intestinal microbiota such as Bacteroidetes and Firmicutes, two phyla significantly altered in IBD (110,195,196). In vitro, the addition of TNF-α to intestinal epithelial cell monolayers results in increased permeability (99).
Further evidence of imbalanced host-microbe interactions in IBD is the ability of increased anti-microbial antibodies in patient serum to predict disease phenotype in CD years before diagnosis (197). Patients with higher antimicrobial antibodies were more likely to have complicated CD, with higher titres predicting earlier complicated disease (197). In pCD, anti-Saccharomyces cerevisiae, anti-outer membrane porin C (an E. coli antigen), and anti-Cbir1 (a flagellin protein), are also predictive of complicated disease (198). The more antimicrobial antibodies a child with pCD was positive for, the higher the likelihood of an earlier progression to complicated disease and need for surgical intervention (198). Antimicrobial antibodies are generally less common in pUC (69), but associations with faecal calprotectin (a marker of mucosal inflammation) levels in pUC have been found (199). The reduced associations between antimicrobial antibodies in pUC compared with pCD may be reflective of the degree of dysbiosis, with the CD microbiome typically further distinguished from healthy controls than the UC microbiome (107).
Treatments in pIBD: links to microbes and diet
Traditional therapies
Despite the described advances in understanding the pathogenesis of IBD, most current treatments for IBD focus on reducing inflammation, mainly through suppression of the immune system. Treatment of pIBD has some variations between pCD and pUC, but both can include glucocorticoids (GCS), thiopurines, methotrexate, aminosalicylates (5-ASA), and biologic therapy (200,201). Dietary therapy, which is unique in that it does not suppress the immune system, is a recommended therapy currently specific to pCD, with exclusive enteral nutrition (EEN) recognized as the first line therapy for luminal pCD (discussed in detail below) (201). 5-ASAs are anti-inflammatory drugs and are suggested as a first-line therapy to induce and maintain remission in mild or moderate pUC, and GCS are the second choice for therapy to induce remission in both pCD and pUC (200,201). GCS carry the risk of steroid-dependence, and have many side effects including increased risk for infections, diabetes, osteoporosis, and cataracts (202). Biologic therapy, usually anti-TNF, is recommended for chronically active or steroid dependent/resistant pUC when 5-ASA and thiopurines do not control disease, and is used for both induction and maintenance of remission (200). Biologics are compounds purified from another organism or virus (203). In pUC, FMT and antibiotics are not currently recommended; but probiotics like VSL#3 (a cocktail of several Lactobacillus, Bifidobacterium and Streptococcus salivarius subsp. thermophilus) or E. coli Nissle 1917 are approved as adjuvant therapy in mild pUC with limited effect. Well powered RCT studies are still needed to prove if probiotics are more effective than placebo (204).
Effects of therapy on host-microbe interactions remain poorly elucidated, even among therapies targeting the microbiota. Intestinal microbiome-altering effects of GCS in animals in both normal and inflammatory conditions include increased Bifidobacterium and Lactobacillus species with elimination of mucin degrader Mucispirillum (204). In pUC patients, GCS use was associated with increased Actinomyces abundance in patients who sustained remission, with an accompanying decrease in a Clostridium OTU (111). Thiopurines may increase mucosal bacteria numbers and adherence, decrease faecal bacterial diversity and richness, and in vitro inhibit Mycobacterium avium subspecies paratuberculosis (204), which is thought by some to play a causative role in CD due to disease similarities with mycobacterial enteritis (205). FMT, most commonly used in recurrent CDI, is being investigated for utility in IBD and shows some promise in UC and CD, albeit at a lower success rate than in CDI (187). Complexity in the host-microbe relationship of IBD when compared to CDI may explain the mixed results and weaker relationships so far observed between FMT and remission in IBD (204). As described above, understanding of the unique microbial dysbiosis present in each patient may be required to adequately modify the microbiome therapeutically (Figure 1).
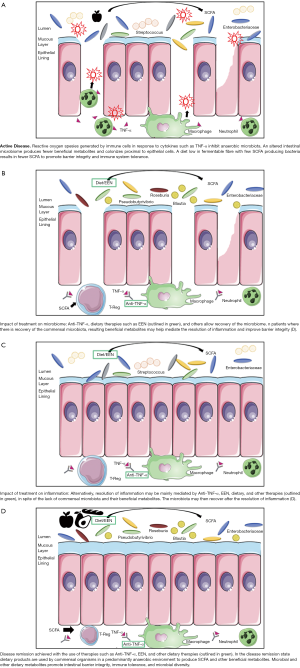
Anti-TNF therapy
Anti-TNF-α therapy is recommended in pCD when, despite immunomodulatory therapy, the patient has persistent active luminal disease, active steroid-refractory disease, active perianal fistulizing disease, and with severe disease at presentation with growth retardation (201). In pUC, anti-TNF-α therapy is recommended for chronically active or steroid-dependent disease that is not controlled by 5-ASA and thiopurines (200). IFX and Adalimumab (ADA) are monoclonal antibodies (MAbs) that can recognize both membrane-bound and secreted TNF-α (206). Membrane-bound TNF may play more of a role than secreted, soluble TNF; agents that preferentially block soluble TNF have not been shown effective for IBD therapy (193). Blocking of TNF effects by IFX results in T-cell apoptosis and reduction in cytokines, chemokines, and adhesion molecules that promote the influx of immune cells into the gut (207). However, the relationship between TNF-α and mucosal healing is complex, as TNF-α knockout mice are more likely to develop colitis with DSS exposure (99). Furthermore, the ability of the probiotic VSL#3 to decrease intestinal permeability in inflamed ileal tissue ex vivo was shown to be TNF dependent in mice (208). Regardless, in vivo anti-TNF- therapy in pIBD patients has been shown to improve the intestinal epithelial barrier and promote mucosal healing (209,210). Risks and side-effects of anti-TNF-α include an increased risk of infection, paradoxical psoriasis in pCD, and infusion reactions (16). Infusion reactions can include the development of antibodies against IFX or ADA, which has been correlated with worsened drug effectiveness (211).
After eight weeks of anti-TNF therapy in pCD patients, faecal microbiota of treated patients more closely resembled healthy controls (65). pCD patients with active disease were found to have reduced relative abundance of almost one third of genera compared to healthy controls, including: Roseburia, Ruminococcus, Akkermansia, Prevotella, Coprococcus, and Eubacterium (65). Shannon diversity and OTU numbers increase with IFX therapy in pCD, with fewer differences in microbiota taxa comparing pCD and controls after IFX treatment than at baseline (212). Wang et al. found that pCD patients who would have a sustained response to IFX therapy (defined here as a PCDAI score of ≤10 throughout follow-up), also showed increased levels of SCFA-producing genera with IFX therapy, including: Blautia, Faecalibacterium, Lachnospira, and Roseburia (158). It remains to be seen, with these consistent changes in microbiota with anti-TNF-α therapy, if these shifts are a cause or consequence of therapy success. Because IFX therapy is administered via intravenous infusion, and ADA subcutaneously, there is no direct interaction with the host intestinal microbiome. Resultant microbial changes may be due to modified host-microbe interactions in the form of decreased inflammation through mediators such as reactive oxygen species or antimicrobial peptides, or shifts may reflect other changes concomitant with therapy such as diet.
Dietary therapy
EEN therapy is currently the first line therapy for induction of remission in pCD and entails liquid meal replacement beverage and the exclusion of all other food and beverages except water for a period of typically six-eight weeks (213-215). The EEN formulas most commonly used include Modulen IBD®, Ensure/Ensure Plus®, Pediasure®, or Nutren/Nutren Junior®; formulas can be elemental or polymeric (216). Although the safety of this treatment is excellent, with no concern for side effects, EEN poses considerable challenges to patients. The most commonly described challenges involve the palatability and monotony of the diet, as well as cost (216). The exact mechanism of action of EEN remains unknown, but theories include immune and/or microbial modulation, removal of many dietary antigens and other ‘offending foods’ (such as preservatives and emulsifiers), and improved intestinal barrier function with decreased inflammation, as reviewed more specifically by others (163,213,215).
EEN therapy significantly impacts the microbiome within one week of initiation (65), and preliminary studies indicate differential changes in the microbiota of responders vs. non-responders to EEN. When comparing newly diagnosed pUC and pCD patients receiving either EEN or corticosteroids for induction therapy, achieving remission was found to better predict changes in patient microbiota than either the therapy or disease subtype (217). Kaakoush et al. found that responders to EEN had a greater decrease in OTUs than non-responders (response was defined as a PCDAI <10 after eight to 12 weeks of therapy) (218). Leach et al. found that patients with the greatest decreases in Bacteroides and Prevotella species with EEN treatment demonstrated improvement in PCDAI, with microbiota changes persisting even four months after EEN completion (219). In a study by Quince et al., EEN resulted in decreases in Bifidobacterium, Ruminococcus, and Faecalibacterium; Shannon diversity also decreased on EEN but returned to pre-treatment levels two months after EEN completion (220). These decreases in OTUs and diversity may reflect the lack of fibre in this liquid diet (219,221); decreasing numbers of dysbiosis-associated taxa present at baseline may be an important part of the therapy mechanism or a side-effect of the homogenous diet. Responsiveness of baseline microbes to this drastic change in diet may be a key determinant of therapy response (222).
Dietary therapies are rapidly expanding in pCD, with some in trial for pUC, but many of the microbial shifts associated with successful therapy to induce remission are consistent across the effective dietary therapies. CD-TREAT, another novel dietary therapy for pCD that attempts to recreate EEN dietary exclusions with food intake, induces similar microbial shifts when compared with EEN (223). The Crohn’s Disease Exclusion diet (CDED), which includes whole foods such as fruits, vegetables, carbohydrates, and meats, induced microbial shifts that include decreased Bifidobacterium and Prevotella, similar to EEN, and increases in Roseburia, which is typically decreased in pIBD and is a known butyrate producer (65,158,224). Both EEN and CDED induce rapid response in >80% of pCD cases, seen within 3 weeks (225). It still remains unclear if these microbial shifts are in part responsible for the effectiveness of therapy, or result as a consequence of successful therapy and disease remission.
Expanding horizons for dietary therapy
Dietary interventions remain a safe treatment option for pCD patients; but much of the diet-host-microbe interactions remain poorly understood. Dietary fibres and proteins play an obvious role in this relationship as host microbes are responsible for the fermentation and breakdown of these components into some of the beneficial products discussed above (62,226-228). Williams et al. provide an in-depth review of the health-associated benefits of these products of fermentation (229). Research suggests that typical Western diets, where incidence of IBD is climbing most significantly (2), consists of only 10–20 g of dietary fibre daily, rather than the recommended 26–38 g, limiting the sources of carbon and energy for many intestinal microbes (230). More complexity is added when we closely examine the group of individual dietary fibres which have grown to encompass non-starch polysaccharides (e.g., cellulose, pectin), non-digestible oligosaccharides (e.g., fructooligosaccharides, galato-oligosaccharides), non-carbohydrate-based polymers (e.g., polyphenols, lignan), and carbohydrates considered to be of animal origin (e.g., chitin) (231).
Dietary fibres are commonly categorized based on their water solubility factor where soluble fibres (pectin, arabinoxylan, β-glucans, inulin, fructo-oligosaccharides, galacto-oligosaccharides, and xyloglucans) are more readily available as a source of nutrition for microbes than their non-soluble counterparts (cellulose and lignin) (61,229,232). Microbial fermentation of dietary fibres involves a variety of species working in a coordinated community where each microbe plays a key role in the fermentation pathway, relying on partner organisms to complete the process (233-236). Many patients experience worsened symptoms following consumption of dietary fibres, and as discussed above, dysbiosis is a well-supported hallmark of IBD; it is possible that production of beneficial products of fibre fermentation are altered or reduced in IBD patients (118,237-240). This suggests that precision tailoring of fibre intake in IBD patients to ensure the capability of fermentation by their unique microbiomes may help to promote healthy levels of beneficial fibre fermentation products. Although some clinicians have promoted reduced fibre consumption, or fibre avoidance, in these patients, there is currently no evidence to support this as a broad therapy in IBD (241,242). When microbes capable of fermentation are present, fibre supplementation may have beneficial effects in IBD; animal models of IBD have demonstrated reduced C-reactive protein (CRP) levels and amelioration of disease activity and histology scores when given a probiotic/fibre-based prebiotic compared to prebiotic alone (243). Similar results have been shown in human studies of prebiotics in IBD patients (244,245). Currently, however, our understanding of the precise role of select fibre types in IBD remains poorly understood.
The hemicellulose fibre arabinoxylan (AX) is a large component of the fibre found in our diets, and is sourced from cereal grains like rye, wheat, oats, barley, rice, sorghum, and some legumes (246-251). A number of microbes ferment AX to SCFA such as acetate, propionate, butyrate, and ethanol (249,252-254). Studies of AX have demonstrated a protective role through stimulation of the caecal mucin layer, and reduced inflammatory markers in clinical trials where adult UC was ameliorated with no significant side effects (255-259). Lactobacilli, Enterococcus, and Bifidobacteria can ferment β-glucan fibres that are found in a variety of plant and fungal cell walls, including mushrooms, oats, and barley (260-263). The effects of β-glucan intake on IBD histological scores is highly dependent on the fibre source, with the most effective source to date from the mushroom species Pleurotus eryngii (264). Similarly, β-fructans, which are commonly found in roots, artichokes, banana, wheat, onion, and garlic are fermented primarily by acetate-producing and acetate-converting butyrate-producing strict anaerobic bacteria, which can lead to anti-inflammatory effects (265-267). The fibre pectin has received less attention as an anti-inflammatory therapeutic dietary fibre due to the variability in esterification and therefore by-product production, but is found in a wide variety of fruits and vegetables, and is easily accessible for fermentation by a number of microbes (Bacteroides, Prevotella, Bacillus, Agrobacterium, Pseudomonas, Ralstonia, Dickeya, and yeast) (268-276). Pectin is almost completely fermented to a variety of SCFAs (268-276). Although animal models have demonstrated beneficial effects with SCFA production from cellulose fermentation (277), succinate has been associated with promoting inflammation and bacterial invasion, suggesting a possible mechanism for patient described sensitivity to consumption of dietary fibres (140,141,278-280).
The unique interactions between the host microbiome and dietary fibres in each pIBD patient may help explain some of the mechanisms of the various dietary therapies, as well as highlight a potential avenue for future developments and dietary modifications. Appreciation of the current and desired diet-host-microbe interface in pIBD could allow better tailoring and development of modifying therapies. Novel approaches, building on these principles, would allow incorporation of a patient’s baseline microbiome/metabolome status in the consideration of therapy choice. For example, if specific SCFAs are reduced, microbes responsible for producing them could be added, or prebiotic- rich diets that would promote growth of these recommended microbes.
Conclusion and future perspective
As the global prevalence of pIBD increases, improved understanding of the pathogenesis of these diseases is warranted to prevent poor outcomes in these patients. The associated genetics, immune system changes, diet, and microbiome suggest that an altered-host microbe relationship is important to development and perpetuation of disease. Current and novel therapies modify this relationship significantly, and in some cases improving this interface may be integral to achieving remission. Evaluation of the baseline host-microbe relationship in patients poses a novel arena for therapy development and dietary modification, as the metabolic capabilities of the patient’s microbiome may impact a patient’s response to therapy. As ongoing work clarifies this diet-host-microbe relationship, considering baseline microbiome status, targeting members of the microbiota and their capabilities through diet and other therapies may improve rates of disease remission and outcomes for paediatric patients with IBD.
Acknowledgments
Funding: SD was supported by student awards from Canadian Institutes of Health Research (CIHR); the Women & Children’s Health Research Institute; and the University of Alberta. HA was supported by a CIHR Fellowship. The Wine lab was funded by CIHR and the Weston Family Foundation.
Footnote
Reporting Checklist: The authors have completed the narrative review checklist. Available at http://dx.doi.org/10.21037/dmr-20-160
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/dmr-20-160). Dr. Wine reports personal fees from AbbVie, Janssen, Nestle Health Sciences and Mead Johnson Nutrition, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2017;390:2769-78. [Crossref] [PubMed]
- Coward S, Clement F, Benchimol EI, et al. Past and Future Burden of Inflammatory Bowel Diseases Based on Modeling of Population-Based Data. Gastroenterology 2019;156:1345-53.e4. [Crossref] [PubMed]
- Roberts SE, Thorne K, Thapar N, et al. A systematic review and meta-analysis of paediatric inflammatory bowel disease incidence and prevalence across Europe. J Crohns Colitis 2020;14:1119-48. [Crossref] [PubMed]
- Kelsen J, Baldassano RN. Inflammatory bowel disease: the difference between children and adults. Inflamm Bowel Dis 2008;14:S9-S11. [Crossref] [PubMed]
- Ye Y, Manne S, Treem WR, et al. Prevalence of inflammatory bowel disease in pediatric and adult populations: recent estimates from large national databases in the United States, 2007–2016. Inflamm Bowel Dis 2020;26:619-25. [PubMed]
- Shaw KA, Cutler DJ, Okou D, et al. Genetic variants and pathways implicated in a pediatric inflammatory bowel disease cohort. Genes Immun 2019;20:131-42. [Crossref] [PubMed]
- Salvador-Martín S, Bossacoma F, Pujol-Muncunill G, et al. Genetic Predictors of Long-term Response to Antitumor Necrosis Factor Agents in Pediatric Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr 2020;71:508-15. [Crossref] [PubMed]
- Lin Z, Wang Z, Hegarty JP, et al. Genetic association and epistatic interaction of the interleukin-10 signaling pathway in pediatric inflammatory bowel disease. World J Gastroenterol 2017;23:4897. [Crossref] [PubMed]
- Penagini F, Dilillo D, Borsani B, et al. Nutrition in pediatric inflammatory bowel disease: from etiology to treatment. A systematic review. Nutrients 2016;8:334. [Crossref] [PubMed]
- Strisciuglio C, Giugliano F, Martinelli M, et al. Impact of environmental and familial factors in a cohort of pediatric patients with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2017;64:569-74. [Crossref] [PubMed]
- Vitale A, Strisciuglio C, Vitale S, et al. Increased frequency of regulatory T cells in pediatric inflammatory bowel disease at diagnosis: a compensative role? Pediatr Res 2020;87:853-61. [Crossref] [PubMed]
- Aujnarain A, Mack DR, Benchimol EI. The role of the environment in the development of pediatric inflammatory bowel disease. Curr Gastroenterol Rep 2013;15:326. [Crossref] [PubMed]
- Kolho K-L, Korpela K, Jaakkola T, et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol 2015;110:921-30. [Crossref] [PubMed]
- Wright EK, Kamm MA, Teo SM, et al. Recent advances in characterizing the gastrointestinal microbiome in Crohn's disease: a systematic review. Inflamm Bowel Dis 2015;21:1219-28. [PubMed]
- Mamula P, Telega GW, Markowitz JE, et al. Inflammatory bowel disease in children 5 years of age and younger. Am J Gastroenterol 2002;97:2005-10. [Crossref] [PubMed]
- Corica D, Romano C. Biological therapy in pediatric inflammatory bowel disease. J Clin Gastroenterol 2017;51:100-10. [Crossref] [PubMed]
- Wong K, Isaac DM, Wine E. Growth Delay in Inflammatory Bowel Diseases: Significance, Causes, and Management. Dig Dis Sci 2021;66:954-64. [Crossref] [PubMed]
- MacKner LM, Crandall WV, Szigethy EM. Psychosocial functioning in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2006;12:239-44. [Crossref] [PubMed]
- Polito JM, Childs B, Mellits ED, et al. Crohn's disease: Influence of age at diagnosis on site and clinical type of disease. Gastroenterology 1996;111:580-6. [Crossref] [PubMed]
- Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 2011;17:1314-21. [Crossref] [PubMed]
- Vernier–Massouille G, Balde M, Salleron J, et al. Natural history of pediatric Crohn's disease: a population-based cohort study. Gastroenterology 2008;135:1106-13. [Crossref] [PubMed]
- Guariso G, Gasparetto M, Dalla Pozza LV, et al. Inflammatory bowel disease developing in paediatric and adult age. J Pediatr Gastroenterol Nutr 2010;51:698-707. [Crossref] [PubMed]
- Kim BJ, Song SM, Kim KM, et al. Characteristics and trends in the incidence of inflammatory bowel disease in Korean children: a single-center experience. Dig Dis Sci 2010;55:1989-95. [Crossref] [PubMed]
- Sawczenko A, Sandhu BK. Presenting features of inflammatory bowel disease in Great Britain and Ireland. Arch Dis Child 2003;88:995-1000. [Crossref] [PubMed]
- Martins AL, Volpato RA, da Penha Zago-Gomes M. The prevalence and phenotype in Brazilian patients with inflammatory bowel disease. BMC Gastroenterol 2018;18:87. [Crossref] [PubMed]
- Zeng Z, Zhu Z, Yang Y, et al. Incidence and clinical characteristics of inflammatory bowel disease in a developed region of Guangdong Province, China: A prospective population-based study. J Gastroenterol Hepatol 2013;28:1148-53. [Crossref] [PubMed]
- Frolkis AD, Dykeman J, Negrón ME, et al. Risk of surgery for inflammatory bowel diseases has decreased over time: a systematic review and meta-analysis of population-based studies. Gastroenterology 2013;145:996-1006. [Crossref] [PubMed]
- Ross SC, Strachan J, Russell RK, et al. Psychosocial functioning and health-related quality of life in paediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2011;53:480-8. [Crossref] [PubMed]
- Malham M, Jakobsen C, Paerregaard A, et al. The incidence of cancer and mortality in paediatric onset inflammatory bowel disease in Denmark and Finland during a 23-year period: a population-based study. Aliment Pharmacol Ther 2019;50:33-9. [Crossref] [PubMed]
- Kugathasan S, Judd RH, Hoffmann RG, et al. Epidemiologic and clinical characteristics of children with newly diagnosed inflammatory bowel disease in Wisconsin: a statewide population-based study. J Pediatr 2003;143:525-31. [Crossref] [PubMed]
- Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527-39. [Crossref] [PubMed]
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307-17. [Crossref] [PubMed]
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut 2013;62:1505-10. [Crossref] [PubMed]
- Cleynen I, González JR, Figueroa C, et al. Genetic factors conferring an increased susceptibility to develop Crohn's disease also influence disease phenotype: results from the IBDchip European Project. Gut 2013;62:1556-65. [Crossref] [PubMed]
- Al Nabhani Z, Dietrich G, Hugot JP, et al. Nod2: The intestinal gate keeper. PLoS Pathog 2017;13:e1006177 [Crossref] [PubMed]
- Bonen DK, Ogura Y, Nicolae DL, et al. Crohn's disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology 2003;124:140-6. [Crossref] [PubMed]
- Tan G, Zeng B, Zhi F-C. Regulation of human enteric α-defensins by NOD2 in the Paneth cell lineage. Eur J Cell Biol 2015;94:60-6. [Crossref] [PubMed]
- Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001;411:603-6. [Crossref] [PubMed]
- Hisamatsu T, Suzuki M, Reinecker H-C, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 2003;124:993-1000. [Crossref] [PubMed]
- Schäffler H, Geiss D, Gittel N, et al. Mutations in the NOD2 gene are associated with a specific phenotype and lower anti-tumor necrosis factor trough levels in Crohn's disease. J Dig Dis 2018;19:678-84. [Crossref] [PubMed]
- Wine E, Reif SS, Leshinsky-Silver E, et al. Pediatric Crohn's disease and growth retardation: the role of genotype, phenotype, and disease severity. Pediatrics 2004;114:1281-6. [Crossref] [PubMed]
- Leshchiner ES, Rush JS, Durney MA, et al. Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease. Proc Natl Acad Sci U S A 2017;114:11392-7. [Crossref] [PubMed]
- Hsu Y-MS, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol 2007;8:198-205. [Crossref] [PubMed]
- Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 2019;25:377-88.e6. [Crossref] [PubMed]
- Prager M, Büttner J, Haas V, et al. The JAK2 variant rs10758669 in Crohn’s disease: altering the intestinal barrier as one mechanism of action. Int J Colorectal Dis 2012;27:565-73. [Crossref] [PubMed]
- Hedl M, Proctor DD, Abraham C. JAK2 disease-risk variants are gain of function and JAK signaling threshold determines innate receptor-induced proinflammatory cytokine secretion in macrophages. J Immunol 2016;197:3695-704. [Crossref] [PubMed]
- Ghoreschi K, Jesson MI, Li X, et al. Modulation of innate and adaptive immune responses by tofacitinib (CP-690,550). J Immunol 2011;186:4234-43. [Crossref] [PubMed]
- Feagan B. Update on tofacitinib for inflammatory bowel disease. Gastroenterol Hepatol 2016;12:572. [PubMed]
- Vancamelbeke M, Vanuytsel T, Farré R, et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm Bowel Dis 2017;23:1718-29. [Crossref] [PubMed]
- Darsigny M, Babeu JP, Dupuis AA, et al. Loss of hepatocyte-nuclear-factor-4α affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One 2009;4:e7609 [Crossref] [PubMed]
- Mohanan V, Nakata T, Desch AN, et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 2018;359:1161-6. [Crossref] [PubMed]
- Manzanillo P, Mouchess M, Ota N, et al. Inflammatory bowel disease susceptibility gene C1ORF106 regulates intestinal epithelial permeability. ImmunoHorizons 2018;2:164-71. [Crossref] [PubMed]
- Fujimoto K, Kinoshita M, Tanaka H, et al. Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol 2017;10:446-59. [Crossref] [PubMed]
- Chang J, Leong RW, Wasinger VC, et al. Impaired intestinal permeability contributes to ongoing bowel symptoms in patients with inflammatory bowel disease and mucosal healing. Gastroenterology 2017;153:723-31.e1. [Crossref] [PubMed]
- Kiesslich R, Duckworth C, Moussata D, et al. Local barrier dysfunction identified by confocal laser endomicroscopy predicts relapse in inflammatory bowel disease. Gut 2012;61:1146-53. [Crossref] [PubMed]
- Martini E, Krug SM, Siegmund B, et al. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol 2017;4:33-46. [Crossref] [PubMed]
- Peters LA, Perrigoue J, Mortha A, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49:1437. [Crossref] [PubMed]
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016;14:e1002533 [Crossref] [PubMed]
- Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol 2015;31:69. [Crossref] [PubMed]
- Kamada N, Núñez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 2014;146:1477-88. [Crossref] [PubMed]
- Armstrong H, Mander I, Zhang Z, et al. Not all fibres are born equal; variable response to dietary fibre subtypes in IBD. Front Pediatr 2021;8:620189 [Crossref] [PubMed]
- Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014;146:1449-58. [Crossref] [PubMed]
- Chehoud C, Albenberg LG, Judge C, et al. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis 2015;21:1948-56. [Crossref] [PubMed]
- Mukhopadhya I, Hansen R, Meharg C, et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect 2015;17:304-10. [Crossref] [PubMed]
- Lewis James D, Chen Eric Z, Baldassano Robert N, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015;18:489-500. [Crossref] [PubMed]
- Imai T, Inoue R, Kawada Y, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol 2019;54:149-59. [Crossref] [PubMed]
- Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039-48. [Crossref] [PubMed]
- Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 2008;43:831-41. [Crossref] [PubMed]
- Birimberg-Schwartz L, Wilson DC, Kolho KL, et al. pANCA and ASCA in children with IBD-unclassified, Crohn's colitis, and ulcerative colitis—a longitudinal report from the IBD Porto Group of ESPGHAN. Inflamm Bowel Dis 2016;22:1908-14. [Crossref] [PubMed]
- Lam S, Zuo T, Ho M, et al. fungal alterations in inflammatory bowel diseases. Aliment Pharmacol Ther 2019;50:1159-71. [Crossref] [PubMed]
- Liang G, Conrad MA, Kelsen JR, et al. Dynamics of the Stool Virome in Very Early-Onset Inflammatory Bowel Disease. J Crohns Colitis 2020;14:1600-10. [Crossref] [PubMed]
- Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447-60. [Crossref] [PubMed]
- Bolsega S, Basic M, Smoczek A, et al. Composition of the intestinal microbiota determines the outcome of virus-triggered colitis in mice. Front Immunol 2019;10:1708. [Crossref] [PubMed]
- Wang W, Jovel J, Halloran B, et al. Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm Bowel Dis 2015;21:1419-27. [Crossref] [PubMed]
- Babickova J, Gardlik R. Pathological and therapeutic interactions between bacteriophages, microbes and the host in inflammatory bowel disease. World J Gastroenterol: WJG 2015;21:11321. [Crossref] [PubMed]
- Clooney AG, Sutton TD, Shkoporov AN, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019;26:764-78.e5. [Crossref] [PubMed]
- Ungaro F, Massimino L, D’Alessio S, et al. The gut virome in inflammatory bowel disease pathogenesis: From metagenomics to novel therapeutic approaches. United European Gastroenterol J 2019;7:999-1007. [Crossref] [PubMed]
- Lopetuso LR, Ianiro G, Scaldaferri F, et al. Gut virome and inflammatory bowel disease. Inflamm Bowel Dis 2016;22:1708-12. [Crossref] [PubMed]
- Prosberg M, Bendtsen F, Vind I, et al. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol 2016;51:1407-15. [Crossref] [PubMed]
- Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010;139:1844-54.e1. [Crossref] [PubMed]
- Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosaassociated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis 2009;15:653-60. [Crossref] [PubMed]
- Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2008;2:716-27. [Crossref] [PubMed]
- Ng SC, Woodrow S, Patel N, et al. Role of genetic and environmental factors in British twins with inflammatory bowel disease. Inflamm Bowel Dis 2012;18:725-36. [Crossref] [PubMed]
- Virta L, Auvinen A, Helenius H, et al. Association of repeated exposure to antibiotics with the development of pediatric Crohn’s disease—a nationwide, register-based Finnish case-control study. Am J Epidemiol 2012;175:775-84. [Crossref] [PubMed]
- Theochari NA, Stefanopoulos A, Mylonas KS, et al. Antibiotics exposure and risk of inflammatory bowel disease: a systematic review. Scand J Gastroenterol 2018;53:1-7. [Crossref] [PubMed]
- Zuo T, Zhang F, Lui GC, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020;159:944-55.e8. [Crossref] [PubMed]
- Levin AM, Sitarik AR, Havstad SL, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep 2016;6:31775. [Crossref] [PubMed]
- Korpela K, Salonen A, Virta LJ, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016;7:10410. [Crossref] [PubMed]
- Nermes M, Endo A, Aarnio J, et al. Furry pets modulate gut microbiota composition in infants at risk for allergic disease. J Allergy Clin Immunol 2015;136:1688. [Crossref] [PubMed]
- Tun HM, Konya T, Takaro TK, et al. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 2017;5:40. [Crossref] [PubMed]
- Lu K, Cable PH, Abo RP, et al. Gut microbiome perturbations induced by bacterial infection affect arsenic biotransformation. Chem Res Toxicol 2013;26:1893-903. [Crossref] [PubMed]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559-63. [Crossref] [PubMed]
- Johnson AJ, Vangay P, Al-Ghalith GA, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 2019;25:789-802.e5. [Crossref] [PubMed]
- Alam MT, Amos GC, Murphy AR, et al. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog 2020;12:1-8. [Crossref] [PubMed]
- Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382-92. [Crossref] [PubMed]
- Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res 2017;10:63. [Crossref] [PubMed]
- Zhou Y, Zhi F. Lower level of bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-analysis. Biomed Res Int 2016;2016:5828959 [Crossref] [PubMed]
- Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731-6. [Crossref] [PubMed]
- Sánchez de Medina F, Romero-Calvo I, Mascaraque C, et al. Intestinal Inflammation and Mucosal Barrier Function. Inflamm Bowel Dis 2014;20:2394-404. [Crossref] [PubMed]
- Quévrain E, Maubert M, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016;65:415-25. [Crossref] [PubMed]
- Arnal M-E, Lalles J-P. Gut epithelial inducible heat-shock proteins and their modulation by diet and the microbiota. Nutr Rev 2016;74:181-97. [Crossref] [PubMed]
- Xu J, Liang R, Zhang W, et al. Faecalibacterium prausnitzii-derived microbial anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J Diabetes 2020;12:224-36. [Crossref] [PubMed]
- Lallès J-P. Microbiota-host interplay at the gut epithelial level, health and nutrition. J Anim Sci Biotechnol 2016;7:66. [Crossref] [PubMed]
- Hedin CR, van der Gast CJ, Stagg AJ, et al. The gut microbiota of siblings offers insights into microbial pathogenesis of inflammatory bowel disease. Gut Microbes 2017;8:359-65. [Crossref] [PubMed]
- Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011;141:227-36. [Crossref] [PubMed]
- Brand EC, Klaassen MA, Gacesa R, et al. Healthy cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterology 2021; Epub ahead of print. [Crossref] [PubMed]
- Nishino K, Nishida A, Inoue R, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol 2018;53:95-106. [Crossref] [PubMed]
- Shah R, Cope JL, Nagy-Szakal D, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes 2016;7:384-96. [Crossref] [PubMed]
- Fritsch J, Abreu MT. Candida in IBD: Friend or Foe? Cell Host Microbe 2020;27:689-91. [Crossref] [PubMed]
- Michail S, Durbin M, Turner D, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis 2012;18:1799-808. [Crossref] [PubMed]
- Schirmer M, Denson L, Vlamakis H, et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 2018;24:600-10.e4. [Crossref] [PubMed]
- Ryan FJ, Ahern A, Fitzgerald R, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun 2020;11:1512. [Crossref] [PubMed]
- Alipour M, Zaidi D, Valcheva R, et al. Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J Crohns Colitis 2016;10:462-71. [Crossref] [PubMed]
- Roy U, Gálvez EJ, Iljazovic A, et al. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep 2017;21:994-1008. [Crossref] [PubMed]
- Rengarajan S, Vivio EE, Parkes M, et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes 2020;11:405-20. [Crossref] [PubMed]
- Coufal S, Galanova N, Bajer L, et al. Inflammatory bowel disease types differ in markers of inflammation, gut barrier and in specific anti-bacterial response. Cells 2019;8:719. [Crossref] [PubMed]
- Palm NW, De Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000-10. [Crossref] [PubMed]
- Armstrong H, Alipour M, Valcheva R, et al. Host immunoglobulin G selectively identifies pathobionts in pediatric inflammatory bowel diseases. Microbiome 2019;7:1-17. [Crossref] [PubMed]
- Moor K, Diard M, Sellin ME, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017;544:498-502. [Crossref] [PubMed]
- Wilmore JR, Gaudette BT, Atria DG, et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe 2018;23:302-11.e3. [Crossref] [PubMed]
- Castro-Dopico T, Dennison TW, Ferdinand JR, et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 2019;50:1099-114.e10. [Crossref] [PubMed]
- Adams RJ, Heazlewood SP, Gilshenan KS, et al. IgG antibodies against common gut bacteria are more diagnostic for Crohn's disease than IgG against mannan or flagellin. Am J Gastroenterol 2008;103:386-96. [Crossref] [PubMed]
- Lin R, Chen H, Shu W, et al. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med 2018;16:359. [Crossref] [PubMed]
- Torres J, Petralia F, Sato T, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology 2020;159:96-104. [Crossref] [PubMed]
- Zaidi D, Bording-Jorgensen M, Huynh HQ, et al. Increased Epithelial Gap Density in the Noninflamed Duodenum of Children With Inflammatory Bowel Diseases. J Pediatr Gastroenterol Nutr 2016;63:644-50. [Crossref] [PubMed]
- Vivinus-Nebot M, Frin-Mathy G, Bzioueche H, et al. Functional bowel symptoms in quiescent inflammatory bowel diseases: role of epithelial barrier disruption and low-grade inflammation. Gut 2014;63:744-52. [Crossref] [PubMed]
- Hollander D, Vadheim CM, Brettholz E, et al. Increased intestinal permeability in patients with Crohn's disease and their relatives: a possible etiologic factor. Ann Intern Med 1986;105:883-5. [Crossref] [PubMed]
- Munkholm P, Langholz E, Hollander D, et al. Intestinal permeability in patients with Crohn's disease and ulcerative colitis and their first degree relatives. Gut 1994;35:68-72. [Crossref] [PubMed]
- Olson TS, Reuter BK, Scott KG, et al. The primary defect in experimental ileitis originates from a nonhematopoietic source. J Exp Med 2006;203:541-52. [Crossref] [PubMed]
- Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol 2013;14:660. [Crossref] [PubMed]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 2007;19:59-69. [Crossref] [PubMed]
- Armstrong H, Bording-Jorgensen M, Dijk S, et al. The complex interplay between chronic inflammation, the microbiome, and cancer: understanding disease progression and what we can do to prevent it. Cancers 2018;10:83. [Crossref] [PubMed]
- Haverson K, Rehakova Z, Sinkora J, et al. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol 2007;119:243-53. [Crossref] [PubMed]
- Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci 2010;15:25. [Crossref] [PubMed]
- Kozakova H, Schwarzer M, Tuckova L, et al. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell Mol Immunol 2016;13:251-62. [Crossref] [PubMed]
- Schicho R, Shaykhutdinov R, Ngo J, et al. Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J Proteome Res 2012;11:3344-57. [Crossref] [PubMed]
- Stephens NS, Siffledeen J, Su X, et al. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J Crohns Colitis 2013;7:e42-8. [Crossref] [PubMed]
- Kolho KL, Pessia A, Jaakkola T, et al. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis 2017;11:321-34. [PubMed]
- Martin F-P, Ezri J, Cominetti O, et al. Urinary metabolic phenotyping reveals differences in the metabolic status of healthy and inflammatory bowel disease (IBD) children in relation to growth and disease activity. Int J Mol Med Sci 2016;17:1310. [Crossref] [PubMed]
- Zaidi D, Huynh HQ, Carroll MW, et al. Gut Microenvironment and Bacterial Invasion in Paediatric Inflammatory Bowel Diseases. J Pediatr Gastroenterol Nutr 2020;71:624-32. [Crossref] [PubMed]
- Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223-37. [Crossref] [PubMed]
- Martin FP, Su MM, Xie GX, et al. Urinary metabolic insights into host-gut microbial interactions in healthy and IBD children. World J Gastroenterol 2017;23:3643-54. [Crossref] [PubMed]
- Lees HJ, Swann JR, Wilson ID, et al. Hippurate: the natural history of a mammalian–microbial cometabolite. J Proteome Res 2013;12:1527-46. [Crossref] [PubMed]
- Sarosiek I, Schicho R, Blandon P, et al. Urinary metabolites as noninvasive biomarkers of gastrointestinal diseases: A clinical review. World J Gastrointest Oncol 2016;8:459. [Crossref] [PubMed]
- Pallister T, Jackson MA, Martin TC, et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci Rep 2017;7:13670. [Crossref] [PubMed]
- Williams HR, Cox JI, Walker DG, et al. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol 2009;104:1435-44. [Crossref] [PubMed]
- Williams HR, Cox IJ, Walker DG, et al. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn's disease. BMC Gastroenterol 2010;10:108. [Crossref] [PubMed]
- Huda-Faujan N, Abdulamir AS, Fatimah AB, et al. The impact of the level of the intestinal short chain Fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J 2010;4:53-8. [Crossref] [PubMed]
- Treem WR, Ahsan N, Shoup M, et al. Fecal short-chain fatty acids in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 1994;18:159-64. [Crossref] [PubMed]
- Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr 2008;100:297-305. [Crossref] [PubMed]
- Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569-73. [Crossref] [PubMed]
- Venegas DP, Marjorie K, Landskron G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol 2019;10:277. [Crossref] [PubMed]
- Ahmad M, Krishnan S, Ramakrishna B, et al. Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut 2000;46:493-9. [Crossref] [PubMed]
- Agus A, Denizot J, Thevenot J, et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci Rep 2016;6:19032. [Crossref] [PubMed]
- Ferrer-Picón E, Dotti I, Corraliza AM, et al. Intestinal inflammation modulates the epithelial response to butyrate in patients with inflammatory bowel disease. Inflamm Bowel Dis 2020;26:43-55. [Crossref] [PubMed]
- Cavaglieri CR, Nishiyama A, Fernandes LC, et al. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci 2003;73:1683-90. [Crossref] [PubMed]
- Segain JP, Raingeard de la Blétière D, Bourreille A, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn's disease. Gut 2000;47:397-403. [Crossref] [PubMed]
- Wang Y, Gao X, Ghozlane A, et al. Characteristics of Faecal Microbiota in Paediatric Crohn's Disease and Their Dynamic Changes During Infliximab Therapy. J Crohns Colitis 2018;12:337-46. [Crossref] [PubMed]
- Roediger W. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 1982;83:424-9. [Crossref] [PubMed]
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 2001;81:1031-64. [Crossref] [PubMed]
- Pang T, Leach ST, Katz T, et al. Fecal biomarkers of intestinal health and disease in children. Front Pediatr 2014;2:6. [Crossref] [PubMed]
- Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol 2011;106:563-73. [Crossref] [PubMed]
- Levine A, Boneh RS, Wine E. Evolving role of diet in the pathogenesis and treatment of inflammatory bowel diseases. Gut 2018;67:1726-38. [Crossref] [PubMed]
- Niewiadomski O, Studd C, Wilson J, et al. Influence of food and lifestyle on the risk of developing inflammatory bowel disease. Intern Med J 2016;46:669-76. [Crossref] [PubMed]
- Rashvand S, Somi MH, Rashidkhani B, et al. Dietary protein intakes and risk of ulcerative colitis. Med J Islam Repub Iran 2015;29:253. [PubMed]
- Jantchou P, Morois S, Clavel-Chapelon F, et al. Animal protein intake and risk of inflammatory bowel disease: The E3N prospective study. Am J Gastroenterol 2010;105:2195-201. [Crossref] [PubMed]
- Khalili H, de Silva PS, Ananthakrishnan AN, et al. Dietary iron and heme iron consumption, genetic susceptibility, and risk of Crohn's disease and ulcerative colitis. Inflamm Bowel Dis 2017;23:1088-95. [Crossref] [PubMed]
- D'Souza S, Levy E, Mack D, et al. Dietary patterns and risk for Crohn's disease in children. Inflamm Bowel Dis 2008;14:367-73. [Crossref] [PubMed]
- Racine A, Carbonnel F, Chan SS, et al. Dietary patterns and risk of inflammatory bowel disease in Europe: results from the EPIC study. Inflamm Bowel Dis 2016;22:345-54. [Crossref] [PubMed]
- Lo C-H, Lochhead P, Khalili H, et al. Dietary inflammatory potential and risk of Crohn’s disease and ulcerative colitis. Gastroenterology 2020;159:873-83.e1. [Crossref] [PubMed]
- Howe A, Ringus DL, Williams RJ, et al. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. The ISME Journal 2016;10:1217-27. [Crossref] [PubMed]
- Singh RK, Chang H-W, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med 2017;15:73. [Crossref] [PubMed]
- Martinez-Medina M, Denizot J, Dreux N, et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 2014;63:116-24. [Crossref] [PubMed]
- Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: an update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol 2014;5:213. [Crossref] [PubMed]
- Goyal A, Yeh A, Bush BR, et al. Safety, clinical response, and microbiome findings following fecal microbiota transplant in children with inflammatory bowel disease. Inflamm Bowel Dis 2018;24:410-21. [Crossref] [PubMed]
- Caldeira LdF. Fecal microbiota transplantation in inflammatory bowel disease patients: A systematic review and meta-analysis. PLoS One 2020;15:e0238910 [Crossref] [PubMed]
- Bibbò S, Settanni CR, Porcari S, et al. Fecal microbiota transplantation: screening and selection to choose the optimal donor. J Clin Med 2020;9:1757. [Crossref] [PubMed]
- Fang H, Fu L, Wang J. Protocol for fecal microbiota transplantation in inflammatory bowel disease: a systematic review and meta-analysis. Biomed Res Int 2018;2018:8941340 [Crossref] [PubMed]
- Imdad A, Nicholson MR, Tanner-Smith EE, et al. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst Rev 2018;11:CD012774 [Crossref] [PubMed]
- Keshteli A, Millan B, Madsen K. Pretreatment with antibiotics may enhance the efficacy of fecal microbiota transplantation in ulcerative colitis: a meta-analysis. Mucosal Immunol 2017;10:565-6. [Crossref] [PubMed]
- Leonardi I, Paramsothy S, Doron I, et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe 2020;27:823-9.e3. [Crossref] [PubMed]
- Qazi T, Amaratunga T, Barnes EL, et al. The risk of inflammatory bowel disease flares after fecal microbiota transplantation: Systematic review and meta-analysis. Gut Microbes 2017;8:574-88. [Crossref] [PubMed]
- Khanna S, Vazquez-Baeza Y, González A, et al. Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 2017;5:55. [Crossref] [PubMed]
- Hourigan S, Chen L, Grigoryan Z, et al. Microbiome changes associated with sustained eradication of Clostridium difficile after single faecal microbiota transplantation in children with and without inflammatory bowel disease. Aliment Pharmacol Ther 2015;42:741-52. [Crossref] [PubMed]
- Gutin L, Piceno Y, Fadrosh D, et al. Fecal microbiota transplant for Crohn disease: A study evaluating safety, efficacy, and microbiome profile. United European Gastroenterol J 2019;7:807-14. [Crossref] [PubMed]
- Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017;8:238-52. [Crossref] [PubMed]
- Sunkara T, Rawla P, Ofosu A, et al. Fecal microbiota transplant–a new frontier in inflammatory bowel disease. J Inflamm Res 2018;11:321. [Crossref] [PubMed]
- D'Odorico I, Di Bella S, Monticelli J, et al. Role of fecal microbiota transplantation in inflammatory bowel disease. J Dig Dis 2018;19:322-34. [Crossref] [PubMed]
- Allegretti J, Eysenbach LM, El-Nachef N, et al. The current landscape and lessons from fecal microbiota transplantation for inflammatory bowel disease: past, present, and future. Inflamm Bowel Dis 2017;23:1710-7. [Crossref] [PubMed]
- Maxwell EC, Dawany N, Baldassano RN, et al. Diverting ileostomy for the treatment of severe, refractory, pediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2017;65:299-305. [Crossref] [PubMed]
- Keubler LM, Buettner M, Häger C, et al. A multihit model: colitis lessons from the interleukin-10–deficient mouse. Inflamm Bowel Dis 2015;21:1967-75. [Crossref] [PubMed]
- Schaubeck M, Clavel T, Calasan J, et al. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut 2016;65:225-37. [Crossref] [PubMed]
- Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol 2014;14:329-42. [Crossref] [PubMed]
- Rahman MM, McFadden G. Modulation of NF-κB signalling by microbial pathogens. Nat Rev Microbiol 2011;9:291-306. [Crossref] [PubMed]
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011;9:356-68. [Crossref] [PubMed]
- Comito D, Cascio A, Romano C. Microbiota biodiversity in inflammatory bowel disease. Ital J Pediatr 2014;40:32. [Crossref] [PubMed]
- Choung R, Princen F, Stockfisch T, et al. Serologic microbial associated markers can predict Crohn's disease behaviour years before disease diagnosis. Aliment Pharmacol Ther 2016;43:1300-10. [Crossref] [PubMed]
- Dubinsky MC, Kugathasan S, Mei L, et al. Increased immune reactivity predicts aggressive complicating Crohn's disease in children. Clin Gastroenterol Hepatol 2008;6:1105-11. [Crossref] [PubMed]
- Spencer EA, Davis SM, Mack DR, et al. Serologic reactivity reflects clinical expression of ulcerative colitis in children. Inflamm Bowel Dis 2018;24:1335-43. [Crossref] [PubMed]
- Turner D, Ruemmele FM, Orlanski-Meyer E, et al. Management of paediatric ulcerative colitis, part 1: ambulatory care—an evidence-based guideline from European Crohn's and Colitis Organization and European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2018;67:257-91. [Crossref] [PubMed]
- van Rheenen PF, Aloi M, Assa A, et al. The medical management of paediatric Crohn’s disease: an ECCO-ESPGHAN guideline update. J Crohns Colitis. 2020 Oct 7:jjaa161. [Epub ahead of print]. doi:
10.1093/ecco-jcc/jjaa161 . - Saag K, Furst D. Major side effects of systemic glucocorticoids. In: Matteson E, editor. UpToDate. UpToDate, Waltham, MA. (Accessed on May 24, 2020)2020.
- Agency EM. Remicade. 2020. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/remicade. Accessed June 15, 2020 2020.
- Nishida A, Inoue R, Inatomi O, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 2018;11:1-10. [Crossref] [PubMed]
- Naser SA, Ghobrial G, Romero C, et al. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 2004;364:1039-44. [Crossref] [PubMed]
- Danese S. Mechanisms of action of infliximab in inflammatory bowel disease: an anti-inflammatory multitasker. Dig Liver Dis 2008;40:S225-8. [Crossref] [PubMed]
- Pasparakis M. Regulation of tissue homeostasis by NF-κB signalling: implications for inflammatory diseases. Nat Rev Immunol 2009;9:778-88. [Crossref] [PubMed]
- Corridoni D, Pastorelli L, Mattioli B, et al. Probiotic bacteria regulate intestinal epithelial permeability in experimental ileitis by a TNF-dependent mechanism. PLoS One 2012;7:e42067 [Crossref] [PubMed]
- Nobile S, Gionchetti P, Rizzello F, et al. Mucosal healing in pediatric Crohn’s disease after anti-TNF therapy: a long-term experience at a single center. Eur J Gastroenterol Hepatol 2014;26:458-65. [Crossref] [PubMed]
- Hyams J, Damaraju L, Blank M, et al. Induction and maintenance therapy with infliximab for children with moderate to severe ulcerative colitis. Clin Gastroenterol Hepatol 2012;10:391-9.e1. [Crossref] [PubMed]
- Ungar B, Glidai Y, Yavzori M, et al. Association between infliximab drug and antibody levels and therapy outcome in pediatric inflammatory bowel diseases. J Pediatr Gastroenterol Nutr 2018;67:507-12. [Crossref] [PubMed]
- Kowalska-Duplaga K, Kapusta P, Gosiewski T, et al. Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease. J Clin Med 2020;9:687. [Crossref] [PubMed]
- Levine A, Wine E. Effects of enteral nutrition on Crohn’s disease: clues to the impact of diet on disease pathogenesis. Inflamm Bowel Dis 2013;19:1322-9. [Crossref] [PubMed]
- Levine A, Rhodes JM, Lindsay JO, et al. Dietary Guidance from the International Organization for the Study of Inflammatory Bowel Disease. Clin Gastroenterol Hepatol 2020;18:1381-92. [Crossref] [PubMed]
- Ashton JJ, Gavin J, Beattie RM. Exclusive enteral nutrition in Crohn's disease: Evidence and practicalities. Clin Nutr 2019;38:80-9. [Crossref] [PubMed]
- Lawley M, Wu JW, Navas-López VM, et al. Global variation in use of enteral nutrition for pediatric Crohn disease. J Pediatr Gastroenterol Nutr 2018;67:e22-9. [Crossref] [PubMed]
- Hart L, Farbod Y, Szamosi JC, et al. Effect of Exclusive Enteral Nutrition and Corticosteroid Induction Therapy on the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Nutrients 2020;12:1691. [Crossref] [PubMed]
- Kaakoush NO, Day AS, Leach ST, et al. Effect of exclusive enteral nutrition on the microbiota of children with newly diagnosed Crohn's disease. Clin Transl Gastroenterol 2015;6:e71 [Crossref] [PubMed]
- Leach S, Mitchell H, Eng W, et al. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn’s disease. Aliment Pharmacol Ther 2008;28:724-33. [Crossref] [PubMed]
- Quince C, Ijaz UZ, Loman N, et al. Extensive Modulation of the Fecal Metagenome in Children With Crohn's Disease During Exclusive Enteral Nutrition. Am J Gastroenterol 2015;110:1718-30. [Crossref] [PubMed]
- Gerasimidis K, Bertz M, Hanske L, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm Bowel Dis 2014;20:861-71. [Crossref] [PubMed]
- Andoh A, Inoue R, Kawada Y, et al. Elemental diet induces alterations of the gut microbial community in mice. J Clin Biochem Nutr 2019;65:118-24. [Crossref] [PubMed]
- Svolos V, Hansen R, Nichols B, et al. Treatment of active Crohn’s disease with an ordinary food-based diet that replicates exclusive enteral nutrition. Gastroenterology 2019;156:1354-67.e6. [Crossref] [PubMed]
- Levine A, Wine E, Assa A, et al. Crohn’s disease exclusion diet plus partial enteral nutrition induces sustained remission in a randomized controlled trial. Gastroenterology 2019;157:440-50.e8. [Crossref] [PubMed]
- Boneh RS, Van Limbergen J, Wine E, et al. Dietary Therapies Induce Rapid Response and Remission in Pediatric Patients With Active Crohn’s Disease. Clin Gastroenterol Hepatol 2021;19:752-9. [Crossref] [PubMed]
- Hillman ET, Lu H, Yao T, et al. Microbial ecology along the gastrointestinal tract. Microbes Environ 2017;32:300-13. [Crossref] [PubMed]
- Tuddenham S, Sears CL. The intestinal microbiome and health. Curr Opin Infect Dis 2015;28:464. [Crossref] [PubMed]
- Valcheva R, Dieleman LA. Prebiotics: Definition and protective mechanisms. Best Pract Res Clin Gastroenterol 2016;30:27-37. [Crossref] [PubMed]
- Williams BA, Grant LJ, Gidley MJ, et al. Gut fermentation of dietary fibres: physico-chemistry of plant cell walls and implications for health. Int J Mol Sci 2017;18:2203. [Crossref] [PubMed]
- Stephen AM, Champ MM-J, Cloran SJ, et al. Dietary fibre in Europe: current state of knowledge on definitions, sources, recommendations, intakes and relationships to health. Nutr Res Rev 2017;30:149-90. [Crossref] [PubMed]
- Hamaker BR, Tuncil YE. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol 2014;426:3838-50. [Crossref] [PubMed]
- Cockburn DW, Koropatkin NM. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol 2016;428:3230-52. [Crossref] [PubMed]
- Patnode ML, Beller ZW, Han ND, et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 2019;179:59-73.e13. [Crossref] [PubMed]
- Chen T, Long W, Zhang C, et al. Fiber-utilizing capacity varies in Prevotella-versus Bacteroides-dominated gut microbiota. Sci Rep 2017;7:2594. [Crossref] [PubMed]
- Jeffery IB, Claesson MJ, O'toole PW, et al. Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol 2012;10:591-2. [Crossref] [PubMed]
- Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207. [Crossref] [PubMed]
- Wu GD, Bushmanc FD, Lewis JD. Diet, the human gut microbiota, and IBD. Anaerobe 2013;24:117-20. [Crossref] [PubMed]
- Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 2008;3:417-27. [Crossref] [PubMed]
- Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016;14:20-32. [Crossref] [PubMed]
- Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol 2018;9:2247. [Crossref] [PubMed]
- Brown AC, Rampertab SD, Mullin GE. Existing dietary guidelines for Crohn’s disease and ulcerative colitis. Expert Rev Gastroenterol Hepatol 2011;5:411-25. [Crossref] [PubMed]
- Owczarek D, Rodacki T, Domagała-Rodacka R, et al. Diet and nutritional factors in inflammatory bowel diseases. World J Gastroenterol 2016;22:895. [Crossref] [PubMed]
- Shinde T, Perera AP, Vemuri R, et al. Synbiotic supplementation containing whole plant sugar cane fibre and probiotic spores potentiates protective synergistic effects in mouse model of IBD. Nutrients 2019;11:818. [Crossref] [PubMed]
- Faghfoori Z, Navai L, Shakerhosseini R, et al. Effects of an oral supplementation of germinated barley foodstuff on serum tumour necrosis factor-α, interleukin-6 and-8 in patients with ulcerative colitis. Ann Clin Biochem 2011;48:233-7. [Crossref] [PubMed]
- Fritsch J, Garces L, Quintero MA, et al. Low-fat, High-fiber Diet Reduces Markers of Inflammation and Dysbiosis and Improves Quality of Life in Patients With Ulcerative Colitis. Clin Gastroenterol Hepatol 2020; Epub ahead of print. [Crossref] [PubMed]
- Kiszonas AM, Fuerst EP, Morris CF. Wheat arabinoxylan structure provides insight into function. Cereal Chem 2013;90:387-95. [Crossref]
- Amodo R, Neukom H. Minor constituents of wheat flour: the pentosan. New Approaches to Research on Cereal Carbohydrates 1985:241-51.
- Dornez E, Gebruers K, Delcour JA, et al. Grain-associated xylanases: occurrence, variability, and implications for cereal processing. Trends Food Sci Technol 2009;20:495-510. [Crossref]
- Mendis M, Leclerc E, Simsek S. Arabinoxylan hydrolyzates as immunomodulators in lipopolysaccharide-induced RAW264. 7 macrophages. Food Funct 2016;7:3039-45. [Crossref] [PubMed]
- Lu ZX, Walker KZ, Muir JG, et al. Arabinoxylan fiber, a byproduct of wheat flour processing, reduces the postprandial glucose response in normoglycemic subjects. Am J Clin Nutr 2000;71:1123-8. [Crossref] [PubMed]
- Henry RJ. A comparison of the non-starch carbohydrates in cereal grains. J Sci Food Agric 1985;36:1243-53. [Crossref]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012;489:242-9. [Crossref] [PubMed]
- Cooper-Bribiesca B, Navarro-Ocaña A, Díaz-Ruiz G, et al. Lactic acid fermentation of arabinoxylan from nejayote by Streptococcus infantarius ssp. infantarius 25124 isolated from pozol. Front Microbiol 2018;9:3061. [Crossref] [PubMed]
- Pastell H, Westermann P, Meyer AS, et al. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed fecal microbiota. J Agric Food Chem 2009;57:8598-606. [Crossref] [PubMed]
- Kanauchi O, Mitsuyama K, Homma T, et al. Treatment of ulcerative colitis patients by long-term administration of germinated barley foodstuff: multi-center open trial. Int J Mol Med 2003;12:701-4. [Crossref] [PubMed]
- Van den Abbeele P, Gérard P, Rabot S, et al. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ Microbiol 2011;13:2667-80. [Crossref] [PubMed]
- Neyrinck AM, Possemiers S, Druart C, et al. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One 2011;6:e20944 [Crossref] [PubMed]
- James SL, Christophersen CT, Bird AR, et al. Abnormal fibre usage in UC in remission. Gut 2015;64:562-70. [Crossref] [PubMed]
- Mendis M, Leclerc E, Simsek S. Arabinoxylan hydrolyzates as immunomodulators in Caco-2 and HT-29 colon cancer cell lines. Food Funct 2017;8:220-31. [Crossref] [PubMed]
- Akramienė D, Kondrotas A, Didžiapetrienė J, et al. Effects of ß-glucans on the immune system. Medicina 2007;43:597. [Crossref] [PubMed]
- Lam KL, Keung HY, Ko KC, et al. In vitro fermentation of beta-glucans and other selected carbohydrates by infant fecal inoculum: An evaluation of their potential as prebiotics in infant formula. Bioactive Carbohydrates and Dietary Fibre 2018;14:20-4. [Crossref]
- Zhao J, Cheung PC. Fermentation of β-glucans derived from different sources by bifidobacteria: Evaluation of their bifidogenic effect. J Agric Food Chem 2011;59:5986-92. [Crossref] [PubMed]
- El Khoury D, Cuda C, Luhovyy B, et al. Beta glucan: health benefits in obesity and metabolic syndrome. J Nutr Metab 2012;2012:851362 [Crossref] [PubMed]
- Vetvicka V, Gover O, Hayby H, et al. Immunomodulating Effects Exerted by Glucans Extracted from the King Oyster Culinary-Medicinal Mushroom Pleurotus eryngii (Agaricomycetes) Grown in Substrates Containing Various Concentrations of Olive Mill Waste. Int J Med Mushrooms 2019;21:765-81. [Crossref] [PubMed]
- Mitmesser S, Combs M. Prebiotics: Inulin and other oligosaccharides. The microbiota in gastrointestinal pathophysiology. Elsevier; 2017. p. 201-8.
- Valcheva R, Koleva P, Martínez I, et al. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes 2019;10:334-57. [Crossref] [PubMed]
- Dewulf EM, Cani PD, Claus SP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013;62:1112-21. [Crossref] [PubMed]
- Dongowski G, Lorenz A, Anger H. Degradation of pectins with different degrees of esterification by Bacteroides thetaiotaomicron isolated from human gut flora. Appl Environ Microbiol 2000;66:1321-7. [Crossref] [PubMed]
- Sahasrabudhe NM, Beukema M, Tian L, et al. Dietary fiber pectin directly blocks Toll-like receptor 2–1 and prevents doxorubicin-induced ileitis. Front Immunol 2018;9:383. [Crossref] [PubMed]
- Tian L, Scholte J, Borewicz K, et al. Effects of pectin supplementation on the fermentation patterns of different structural carbohydrates in rats. Mol Nutr Food Res 2016;60:2256-66. [Crossref] [PubMed]
- Luis AS, Briggs J, Zhang X, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 2018;3:210-9. [Crossref] [PubMed]
- Larsen N, Bussolo de Souza C, Krych L, et al. Potential of pectins to beneficially modulate the gut microbiota depends on their structural properties. Front Microbiol 2019;10:223. [Crossref] [PubMed]
- Kaur SJ, Gupta VK. Production of pectinolytic enzymes pectinase and pectin lyase by Bacillus subtilis SAV-21 in solid state fermentation. Ann Microbiol 2017;67:333-42. [Crossref]
- Kuivanen J, Biz A, Richard P. Microbial hexuronate catabolism in biotechnology. AMB Express 2019;9:16. [Crossref] [PubMed]
- Dittoe DK, Barabote RD, Rothrock MJ, et al. Assessment of a Potential Role of Dickeya dadantii DSM 18020 as a Pectinase Producer for Utilization in Poultry Diets Based on in silico Analyses. Front Microbiol 2020;11:751. [Crossref] [PubMed]
- Benoit I, Coutinho PM, Schols HA, et al. Degradation of different pectins by fungi: correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics 2012;13:321. [Crossref] [PubMed]
- Armstrong H, Bording-Jorgensen M, Wine E. The Multifaceted Roles of Diet, Microbes and Metabolites in Cancer. Cancers 2021;13:767. [Crossref] [PubMed]
- Kim Y, Hwang SW, Kim S, et al. Dietary cellulose prevents gut inflammation by modulating lipid metabolism and gut microbiota. Gut Microbes 2020;11:944-61. [Crossref] [PubMed]
- Mills E, O’Neill LA. Succinate: a metabolic signal in inflammation. Trends in cell biology 2014;24:313-20. [Crossref] [PubMed]
- Davis SR, Cousins RJ. Metallothionein expression in animals: a physiological perspective on function. J Nutr 2000;130:1085-8. [Crossref] [PubMed]
Cite this article as: Dijk S, Armstrong H, Valcheva R, Wine E. Paediatric IBD: the host, diet & microbes in pathogenesis & treatment: a narrative review. Dig Med Res 2021;4:6.


