
宿主、饮食和微生物在儿童炎症性肠病的发病和治疗中的叙述性综述
简介:小儿炎症性肠病
炎症性肠病(inflammatory bowel diseases,IBD)的全球发病率一直处于稳步上升阶段,目前为止已超过 0.3% [1]。预计到 2030 年,加拿大等西方国家的IBD发病率将高达 1% [2]。IBD在儿童中的发病率也在逐渐增加 [3],高达25% 的确诊IBD病例发生在儿童人群 [4,5]。IBD 有两种主要亚型,克罗恩病(Crohn disease,CD)和溃疡性结肠炎 (ulcerative colitis,UC)。IBD 是一种慢性肠道炎症,在CD中主要表现为沿胃肠道从口腔到肛门的任何地方发生的跳跃性病变,而在UC中炎症则以连续的方式局限于结肠部位。目前,IBD的病因尚未完全清楚,尚无有效治愈方案。有证据表明,遗传、环境、免疫反应和肠道菌群的相互作用与疾病的发生和发展有关 [6-14]。生长和发育问题是儿童炎症性肠病(paediatric inflammatory bowel diseases,pIBD)的常见表征如发育不良等,也是pIBD的临床诊断依据。 此外,pIBD还可能造成儿童线性生长衰竭、青春期延迟和骨密度峰值降低等风险 [15-17]。患有IBD的儿童还可能面临较大的抑郁和焦虑等社会心理问题 [18]。
儿童克罗恩病(paediatric Crohn disease,pCD)患者的诊断年龄越小,患病情况越复杂 [19,20]。复杂疾病在pCD中被定义为狭窄、穿透性病变,且与较差的预后和增加的手术需求密切相关 [21]。与成人相比,pCD患者发生小肠病变的可能性更大(儿童42%,成人15%) [22]。儿童溃疡性结肠炎(paediatric ulcerative colitis,pUC)患者有约43%-81%的可能性患有广泛性炎症的全结肠炎 [22-24],而成人患病率约为20%-35% [22,25,26]。确诊后的10年内,有近一半的pCD患者和约16%的pUC患者需要接受手术治疗 [27],且患有IBD的儿童的健康相关平均生活质量亦较低 [28]。随访研究发现,儿童期确诊IBD的患者与其成年期患癌风险和死亡率增加有关 [29]。为改善pIBD预后以及预防与儿童早期诊断相关的疾病并发症,需要更好地确定pIBD的发病原因及治疗反应的预测因素。我们根据叙述性综述要点清单呈上以下文章(文章链接https://dmr.amegroups.com/article/view/7168/rc )。
方法
本篇文献综述的目的在于阐明 pIBD 中宿主和微生物的相互作用关系,并描述这些关系是如何受饮食和当前疾病疗法的影响。本文优先考虑最近五年发表的经同行评议的原创性英文研究论文。较早前的论文如果能够提供新颖或重要的相关信息也在本文的考虑范围之内。此外,本文优先考虑人体研究的相关证据,其次是动物研究以及体外研究。
病因学和病理生理学:宿主、微生物和饮食
pIBD的发病机制复杂,涉及遗传和环境因素。为了清楚定义微生物和饮食如何参与pIBD的发病机制,以及明确它们如何在治疗反应中起到贡献作用,我们将概述目前已知的pIBD发病机制,包括有关遗传学、微生物、肠道屏障、代谢物和饮食的详细知识。
炎症性肠病相关基因
尽管遗传因素在pIBD中发挥一定作用,但诊断结果表明大多数患者并无家族病史 [30],表明在pIBD的发病机制中环境因素比遗传因素更加重要。Graham和Xavier就IBD的遗传学发表过一篇详细综述 [31]。相关基因在肠道上皮屏障完整性、微生物识别和清除,以及宿主免疫应答调等方面均有参与 [32,33]。半胱天冬酶募集结构域家族成员15号(caspase recruitment domain family member 15,CARD15)基因,又名核苷酸结合寡聚化结构域2(nucleotide-binding oligomerization domain two,NOD2),与IBD的遗传关联最强,且与CD具有特异相关性 [34]。NOD2是胞质中能够识别细菌肽聚糖的模式识别分子 [35,36],具有 IBD 相关 NOD2 变体的个体肠道抗菌肽(例如α-防御素)含量减少 [37],而最常见的 IBD 相关变体在体外对细菌肽聚糖和脂多糖(lipopolysaccharide,LPS)没有反应 [38],并且防御细胞内侵袭的能力急剧下降 [39]。与携带野生型基因的患者相比,携带这些突变基因患者的抗肿瘤坏死因子-α(tumour necrosis factor-α,TNF-α)单克隆抗体(monoclonal antibody,Mab)的剂量间循环水平更低(下文将讨论),从而可能导致治疗效果降低 [40]。携带NOD2变异的儿童也更有可能出现生长障碍 [41]。另一个与IBD相关的基因变异位于半胱天冬酶募集结构域家族成员9号(caspase recruitment domain family member 9,CARD9)基因,它可以导致细胞对胞内病原体的先天免疫反应受损,并通过增加核因子-κB(nuclear factor-κB,NF-κB)信号传递提升促炎信号(包括TNF-α)[42,43]。携带这些变异基因的个体还会表现出限制性马拉色菌增多,一种可以使动物模型结肠炎恶化的真菌 [44]。IBD相关基因NOD2和CARD9变异患者可能出现无法充分清除微生物的情况,致使微生物在细胞内或近上皮肠道环境中持续存在,从而导致慢性炎症。
IBD中也有与肠道通透性增加相关的遗传变异,如CD中的Janus激酶(Janus kinase 2,JAK2)[45]。JAK2参与细胞因子信号转导 [46],而抑制JAK2可以减少T细胞介导的炎症反应 [47]。JAK抑制剂托法替尼已被证明在部分IBD人群中有效 [48],可能的机制包括降低JAK驱动的肠道通透性,减少腔内环境的抗原易位和T细胞介导的炎症产生。其他与屏障完整性降低相关的遗传关联还包括CD中的前列腺素E受体4 (Prostaglandin E Receptor 4,PTGER4)和UC中的肝细胞核因子4α(Hepatocyte Nuclear Factor 4 Alpha,HNF4A)[49],分别调节链接蛋白/细胞骨架蛋白和对上皮紧密连接起重要作用claudin-15蛋白 [50]。参与细胞粘附和紧密连接的其他IBD相关基因变异包括C1orf106和RNF186 [51-53]。肠屏障功能不佳可能导致抗原暴露增加,并延续促炎免疫反应 [54-56]。尽管在IBD中已经确定了大量的遗传关联,但是迄今为止只有26%的CD患者和19%的UC患者的遗传性可以用遗传学来解释 [57]。此外,遗传关联也无法解释最近发病率的上升趋势 [2],突出了评估环境因素作为IBD病理生理学因素的重要性。
参与炎症性肠病的微生物
人体肠道微生物组由细菌、病毒、古细菌和真核生物组成,其中细菌细胞的数量与人体细胞的数量大致相同 [58]。肠道微生物组的改变被认为在许多人类疾病中都扮演着重要角色。微生物组对健康也至关重要,尤其是在健康的免疫发育,以及膳食纤维和蛋白质发酵,合成某些维生素和神经递质等代谢功能方面都是必不可少的 [59-61]。正常的肠道菌群主要由多个门水平的专性厌氧菌组成,包括厚壁菌门、拟杆菌门、放线菌门、变形菌门和疣微菌门、梭菌门、蓝藻门以及待定候选门TM7 [62]。病毒、真菌和其他真核生物在肠道微生物组中的作用尚未得到充分研究,但有证据表明真菌和病毒的共生失调也存在于儿童 [63-65] 和成人IBD [66,67]。真菌在肠道微生物群中所占的比例相对较小;而相较于对照组,IBD患者中抗真菌抗体明显增加,这引发了对真菌的研究兴趣 [69]。 IBD真菌组的特征表现为真菌多样性降低而总体载量增加,担子菌门与子囊菌门比例增加,而酿酒酵母菌与白色念珠菌比例降低 [44,63,67]。此外,之前提到的携带CARD9变异基因的IBD患者的限制性马拉色菌数量也有所增加 [44,70]。Lam等人最近也针对真菌总数、相对丰度和多样性的变化,以及潜在的相关治疗方法进行了综述分析 [70]。目前有关IBD患者中肠道病毒群的变化则只知更少,但其已被证实可在动物IBD模型中引发结肠炎,并可能影响微生物生态系统中的复杂关系,包括噬菌体对细菌菌群的影响 [75,76]。肠道病毒群的潜在影响在多篇综述中均有详细讨论 [77,78]。
与对照组相比,肠道菌群的改变是IBD的明显特征。相较而言,CD和UC的肠道菌群改变亦有所不同,且差异的显著性与疾病的活跃性相关 [79]。大量双生子研究发现,IBD的菌群失调与宿主遗传因素间可能存在部分独立关系 [80,81]。CD双生子的微生物菌群差异支持环境因素在菌群失调的建立和维持中的影响作用 [82,83]。已知的影响肠道微生物群的环境因素包括抗生素暴露 [84,85]、感染(包括COVID-19)[86]、营养以及吸烟等其他环境暴露 [87-93]。IBD中微生物的变化表现为厚壁菌门和放线菌门中严格厌氧菌的菌种减少,而耐氧的变形菌和拟杆菌增加 [94]。其他变化包括 pCD 中黏膜细菌物种丰富度水平降低且该变化并不总能通过粪便分析发现 [95],粪便微生物群多样性减少 [96], 以及某些黏膜细菌类群的改变,包括肠杆菌科数量的增加和严格厌氧厚壁菌门(如梭菌目)丰度的降低 [95]。在 IBD 中也可以看到梭状杆菌、普拉梭菌和双歧杆菌的含量降低,且相比于UC,CD中的相对丰度下降更多 [79]。IBD中的拟杆菌含量也呈下降态势,且在活跃性IBD中更为明显 [97]。普拉梭菌是一种重要的纤维分解菌,已被证明具有抗炎作用,其缺失对IBD复发具有预示作用 [98]。在 IBD 中减少的普拉梭菌等有益微生物可能通过紧密连接蛋白、诱导型热休克蛋白、免疫调节甚至产生抗炎肽来改善粘膜屏障功能 [99-103]。
通过儿童克罗恩病活动指数(paediatric Crohn disease activity index,PCDAI)的测定,微生物失调与疾病严重程度密切相关 [95]。研究发现,微生物甚至可以预测IBD未来的活跃性。通过分析肠杆菌科、梭杆菌和嗜血杆菌的水平,结合疾病发病年龄和诊断时的PCDAI,可以显著预测未来的PCDAI [95]。与对照组相比,CD 患者的健康兄弟姐妹也表现出肠道微生物组的改变,导致他们罹患IBD的风险增加,进而表明微生物组与IBD的发病机制有因果关系 [104-106]。发病位置也与微生物丰度的变化也有关联,与以结肠/回结肠为主的CD患者相比,以回肠为主的CD患者在肠黏膜标本中有较低丰度的普拉梭菌和较高水平的大肠杆菌 [81]。
虽然UC与未患IBD对照组的微生物差异小于CD [107],但Shah等人发现,患者对初始治疗的反应与某些微生物类群的不同丰度有关 [108]。某些微生物可能会预测甚至介导患者在未来对 pUC 治疗的反应,例如 Shah 等人发现某些梭状芽孢杆菌和拟杆菌 的操作分类单元(operational taxonomic units,OTUs)只能在基线时在 pUC 患者中检测到,而这些患者对治疗没有临床反应 [108]。 同样,在活动期UC患者粪便菌群移植后,念珠菌在基线时的高丰度与临床反应和细菌多样性增加相关 [109]。与对照相比,在严重 pUC患者中,门水平变化包括较低水平的厚壁菌门和较高水平的变形杆菌。在对糖皮质激素治疗无效的儿童中,pUC中较低的微生物丰度更为突出 [110]。此外,UC微生物组的波动与疾病的严重程度和结肠切除术有关 [111],而在病情更严重的患者中发现瘤胃球菌和毛螺菌显著减少,而咽峡炎链球菌丰度增加。
虽然在不同的分类水平上都有与疾病严重程度相关的微生物群改变的详细记录 [94],但目前还没发现任何特定的微生物是与疾病暴发有关的罪魁祸首。对炎症和非炎症肠段的黏膜微生物群进行测序比较,尚未确定与个体炎症相关的特定类群,但确实显示出pUC患者非炎症末端回肠的显著组成变化,这表明局部炎症上游的微生物发生改变 [112,113]。很可能复杂的宿主-微生物相互作用是特定于个体的。Roy等人发现动物模型中引入的微生物失调的特定改变可以导致和引入的微生物群落相关的特有结肠炎表型和病理状态 [114]。未来识别每个 IBD 患者特有的“问题微生物群”的技术可能会涉及识别从患者分离的微生物上发现的免疫球蛋白 (immunoglobulin,Ig) 结合水平。在IBD患者中,免疫球蛋白A(Immunoglobulin A,IgA)和免疫球蛋白G(Immunoglobulin G,IgG)显示出不同的微生物结合,包括UC和CD之间的不同结合 [115,116]。在动物和体外模型中,高水平的免疫球蛋白被膜可用于识别来源于IBD患者炎症诱导细菌,表明该技术可用于识别特定患者体内的致病细菌 [117,118]。粘膜IgA可以帮助减少微生物的入侵 [119]。在一些动物模型中,某些针对肠道微生物群的抗体可以防御细菌性脓毒症 [120]。这些抗体在IBD中的作用尚不清楚,但已被探索是否可以作为对肠道细菌的不适当免疫反应的标志物 [121] ,或是疾病活动性增加导通肠道屏障破坏引起抗原暴露增加的标志物 [115,122,123]。一些针对共生微生物和真菌的抗体如抗酿酒酵母(anti-Saccharomyces cerevisiae antibodies,ASCA)在IBD的诊断和预测中作用有限 [124]。
炎症性肠病中改变的肠道屏障
屏障完整性受损是 IBD 的另一个特征 [125-129]。 pUC 患者的粘液屏障厚度降低,含粘蛋白的杯状细胞减少,细菌定植于比对照组更靠近肠上皮细胞的位置 [113]。同样,与对照组相比,pCD 中的细菌定植也更靠近肠上皮细胞的位置 [113]。共聚焦激光内窥镜检查显示,与对照组相比静脉注射的荧光素通过 pCD 和 pUC 肠上皮屏障的渗漏增加 [125];在成人中,荧光素渗漏增加与疾病复发有关 [55]。正如在患者遗传中提到的,受损的上皮屏障可能导致细菌和抗原易位增加,从而延长慢性炎症的持续性。
肠道微生物群的改变和宿主屏障完整性会给IBD患者带来了许多潜在的后果 [130]。微生物可以通过增加或减少对病原体定植的抵抗力、合成维生素和营养物质,以及训练平衡免疫系统的发展来直接影响健康 [130-132]。平衡的宿主-微生物相互作用对健康至关重要;无菌小鼠在心血管、呼吸系统、胃肠道和免疫系统等许多身体系统中表现出异常 [131]。无菌动物也表现出显著的肠道功能和形态变化 [131]。其中一些异常可以通过特定细菌的定植来纠正,包括通过给无菌动物添加某些乳酸杆菌来改善肠道屏障的完整性 [133-135]。IBD 中不健康的宿主-微生物关系可能会延续或引发疾病,而明确这种关系可以提高我们对这些疾病病因的理解。
代谢物:炎症性肠病中饮食与微生物的纽带
代谢物是新陈代谢所必需的,也是在新陈代谢中形成的,代表着饮食、宿主和微生物因素,可以实时反映肠道微环境。代谢产物的鉴定和定量可以为微生物功能和人体代谢提供具体的评价。除了微生物丰度的改变外,尿液代谢物的变化表明 IBD 微生物组中的微生物功能发生了改变。在患有 IBD 的成人患者中,血清和尿液代谢物可以区分疾病的活动性和缓解性 [10]。尿液代谢物可以区分成人 IBD 患者和健康对照,但区分 CD 和 UC 的能力则所有不同 [136,137],而血清和粪便代谢物能够把pCD 从 pUC 和健康对照中区别出来 [138]。有限数量的研究表明,尿液代谢组学也可以将 pIBD 与健康对照区分开来 [139]。在 pIBD 患者结肠镜检查期间收集的粘膜表面样本的代谢组学将肠内琥珀酸盐与体外细菌入侵联系起来,突出了肠道微环境在 IBD 中的重要性 [140]。 Lavelle 和 Sokol 详细介绍了 IBD 中重要的微生物相关代谢物及其潜在作用 [14]。
在儿童和成人 IBD 中常见的代谢物改变属于代谢通路,表明谷胱甘肽代谢、柠檬酸循环 (citric acid cycle,CAC) 以及重要的微生物活性发生变化 [136,139,142]。 马尿酸盐被定义为共代谢物,因为它是由宿主和微生物代谢共同产生的 [143]; 它在无菌动物中明显缺失 [143],并且在 IBD 中显着减少 [144]。 马尿酸盐与微生物组多样性以及水果和全谷物摄入量的增加呈正相关 [145]。 当排除潜在的饮食影响时,马尿酸盐仍然是 IBD 与对照组的标志物 [146]。 补充马尿酸盐前体苯甲酸钠不会增加 CD 或对照组尿液中的马尿酸盐水平,并且 CD 患者尿液中马尿酸盐水平持续降低,表明缺乏相关微生物代谢功能 [147]。
其他微生物相关代谢物包括SCFA,它们由膳食纤维以及少量的氨基酸通过微生物发酵产生;这些在成人和儿童 IBD 中通常显著降低 [136,137,148,149]。 IBD 患者的粪便中通常含有较少的醋酸、丁酸和丙酸,而乳酸和丙酮酸则增加 [148]。 SCFA 的作用包括抑制肠道通透性和增加有助于预防慢性炎症的调节性 T 细胞的数量 [150,151]。 SCFA 还可以通过与肠道 G 蛋白偶联受体如 GPR109A、GPR43 和 GPR41等结合来激活抗炎信号级联反应 [152]。 SCFA 代谢受损与 IBD 相关;例如,实验用葡聚糖硫酸钠 (dextran sulphate sodium,DSS) 诱导的结肠炎的特征是丁酸氧化减少和葡萄糖氧化增加 [153]。此外,在一项研究中发现,无论疾病活动性如何,与对照组相比,CD患者回肠中GPR43的表达显著减少 [154]。炎症会影响IBD中丁酸盐的反应;IBD患者体外类器官和对照组对丁酸的反应类似,但当加入促炎细胞因子TNF-α时,只有IBD患者类器官对丁酸的吸收减少 [155]。这可以解释为什么外源性 SCFAs 不能始终如一地导致 IBD 治疗的临床改善 [156]。尽管体外、离体和动物研究显示炎症标志物减少和肠道屏障改善,但以往直接用SCFA给药(使用丁酸灌肠剂)的研究结果喜忧参半 [157,158]。
细菌发酵产生的丁酸盐是结肠细胞的主要能量来源 [159];毫无疑问,来自无菌小鼠的结肠细胞具有更少的CAC中间酶 [153]。丁酸盐具有多种生理作用,如在体外减少细菌移位 [99],作为能量来源,以及组蛋白脱乙酰酶(histone deacetylase,HDAC)抑制剂 [153]。一些抗性淀粉(resistant starch,RS)会增加丁酸盐浓度,并且 SCFA 的产生会随着饮食和肠道运送时间而变化 [160]。粪便 SCFA 含量并不总是反映发酵的产量,因为较长的运送时间与丁酸盐的减少有关 [160]。运输时间对SCFA浓度的影响可能是由于在较慢的运输过程中增加了吸收和利用的机会,或者与RS和膳食纤维到大肠的运输时间以及微生物代谢和发酵的时间有关 [160]。许多其他因素也会影响 SCFA 浓度;抗生素会减少短链脂肪酸,而摄入不同的膳食纤维会增加不同数量的SCFA [161]。此外,SCFA 还可以改变肠道微生物群,它们的酸度可以选择更耐酸的细菌 [161]。SCFA可能在患者对治疗的反应中发挥作用;Wang等人发现pIBD患者在英夫利昔(Infliximab,IFX)治疗前产生SCFA的细菌显著减少,治疗后则向更接近健康对照组的SCFA转变,并且产生SCFA的物种的丰度可以预测pIBD患者的持续缓解 [158]。但是,需要注意的是当涉及到个体间差异和缺乏标准化的实验方案的时候,要充分认识到代谢组学的局限性。
环境:关注饮食
低纤维、高加工和高脂肪的西方饮食被认为是最近IBD发病率上升的部分原因,尤其是在西方国家 [162]。虽然已经认识到一些饮食风险因素和关联,但大多数是通过病例对照或队列研究确定的。很难设计和实施能够证明因果关系的实验性饮食研究,而在动物研究中,近似人类饮食和消耗模式也是一种挑战。评估与个别食物或营养素的关联可能会错过获取微量元素和常量营养素相互作用的复杂性。其他复杂性还包括饮食摄入量与文化和遗传的关联,以及许多偏见报告。
饮食与成人和儿童IBD风险的增加和降低都有关联。CD与饮食摄入的关系尤其密切 [163]。与IBD风险增加相关的食物包括快餐 [164] 和增加的特定于动物蛋白的蛋白质摄入量 [163,165-167]。保护因素包括母乳喂养,特别是对于pCD保护因素包括增加蔬菜、水果、鱼、橄榄油、谷物和坚果的摄入 [168]。一些性别特异性关联也已被确认,例如与女性pCD风险增加相关的食物包括肉类、含糖食物和高脂食物 [168]。其中许多研究都是前瞻性的,如E3N [166]、EPIC [169]、护士健康研究I和II以及健康职业人员随访研究 [170],表明饮食摄入可能建立有利于IBD发展的肠道环境。饮食被认为是通过改变宿主免疫、宿主屏障完整性和微生物群(包括病毒)的改变来发挥作用 [163,171]。Levine等人在综述中深入讨论了潜在的机制;我们也将在这里简要地讨论它们 [163]。这些因素中的宿主变异性,特别是微生物群的差异,可能在与某些饮食成分(如肉类或膳食纤维)的复杂关联中发挥重要作用。在缺乏膳食纤维的情况下,一些微生物表现出增强的黏液溶解活性-消化和消耗宿主肠道黏液层,损害肠道屏障的完整性,进而导致免疫激活增加 [163]。由此产生上皮屏障破坏,正如间隙密度增加所证明的那样,与炎症和疾病活动性的增加呈正相关 [125]。膳食纤维和RS可增加SCFA的产生,潜在地降低肠道通透性 [163]。其他饮食成分也可能影响SCFA受体的水平;在高脂高糖饮食的小鼠中,GPR43受体表达减少 [154]。
饮食是肠道微生物组的主要决定因素之一 [172]。早前的研究发现饮食模式对肠道微生物组的预测能力显著优于单一营养成分,这可能是因为天然食品之间的复杂相互作用无法从单独的成分中预测 [93]。在小鼠中,高脂高糖的西式饮食增加了病原微生物的定植,如粘附侵袭性大肠杆菌( adherent invasive E. coli, AIEC) [173],这是和CD病理生理学有关的关键病原微生物 [174]。针对宿主、饮食和微生物之间的这些关联,已经开发了一些饮食疗法,并在下面的治疗部分进行了描述。
炎症性肠病中的饮食-宿主-微生物相互作用
IBD中的遗传、微生物、饮食和代谢关联表明,宿主-微生物关系失调,再加上饮食的进一步影响,是重要的IBD发病机制。有证据表明,以微生物为靶点的治疗,如抗生素和饮食,可导致IBD的临床改善 [99,163]。粪便微生物群移植(faecal microbiota transplant,FMT)是一种新兴的以微生物为基础的治疗方法,已经显示出一些前景,但在儿童方面的治疗经验仍然有限[175]。虽然目前存在各种FMT的方案,但是在许多方面上存在矛盾,例如供体选择(普遍的可能是有益的)[176,177],新鲜供体粪便与冷冻供体粪便 [178],以及是否需要抗生素预处理 [178-180]。基线患者微生物群可能是FMT结果可变性的一个因素;在成人UC中,有证据表明基线真菌菌群,特别是念珠菌丰度的增加,可以预测阳性FMT反应 [181]。许多关于FMT的证据对于治疗叠加性艰难梭菌感染(Clostridioides difficile infection,CDI)是特异性的 [182]。与为患IBD的患者相比,FMT治疗CDI的效果较差,仅有较少的微生物发生转移 [183,184]。尽管IBD中大多数FMT的不良事件是轻微的,包括腹泻、恶心和腹痛等非特异性症状 [176],但是可能会发生一些潜在的严重疾病 [182,185]。最近的几篇综述进一步讨论了IBD中的FMT [179,186-189]。
另一个例子是,pIBD中通过回肠造口转移粪流可达到临床上的改善效果,当粪流返回结肠时疾病严重程度往往再次增加 [190]。此外,几乎所有结肠炎动物模型中,如白细胞介素-10(interleukin-10, IL-10)敲除或TNFΔARE小鼠,在无菌条件下不会发生肠道炎症 [191,192]。
可能是由于遗传和环境/饮食因素,IBD患者腔内抗原通过肠屏障的泄漏增加,导致黏膜组织中过度的细胞因子依赖性炎症反应 [193]。这些抗原包括细菌的产物或成分,如LPS或肽聚糖。微生物抗原可以通过增加TNF-α的生成来增加NF-κB信号通路,这被认为是抵御入侵病原体的重要组成部分 [194]。TNF-α在在IBD中的发病机制主要是通过激活成纤维细胞、增加促炎细胞因子生成以及增强T细胞对细胞凋亡的抵抗力等发挥作用 [193]。TNF-α通过NF-κB的作用也导致Paneth细胞死亡,肠上皮细胞受损,肌成纤维细胞的基质金属蛋白酶(matrix metalloproteinases,MMPs)合成增加 [193]。Paneth细胞是位于小肠肠隐窝的特化分泌肠上皮细胞(在IBD的大肠中也有发现),对产生抗菌和免疫刺激分子非常重要 [195]。它们在抵御肠道病原体和调节某些肠道微生物菌群(如拟杆菌门和厚壁菌门)的丰度方面发挥作用,这两个菌门在IBD中显著变化 [110,195,196]。在体外,在肠上皮细胞单层中加入TNF-α则会导致通透性增加 [99]。
IBD中宿主-微生物相互作用不平衡的进一步证据是患者血清中抗微生物抗体的增加能够在确诊前几年预测CD的疾病表型 [197]。抗微生物抗体较高的患者更有可能患有复杂性CD,而较高的滴度预示着更早出现的复杂疾病 [197]。在pCD中,抗酿酒酵母、抗外膜孔蛋白C(一种大肠杆菌抗原)和抗Cbir1(一种鞭毛蛋白)也可预测复杂疾病 [198]。pCD患儿的抗微生物抗体阳性值越高,早期进展为复杂疾病和需要手术干预的可能性越高 [198]。抗微生物抗体在pUC中通常不太常见 [69],但已发现与pUC中粪钙保护素(粘膜炎症的标志)的水平相关。与pCD相比,pUC中抗微生物抗体之间的相关性降低可能反映了生态失调的程度,且与UC微生物组相比,CD微生物组通常与健康对照组有更多的区别 [107]。
儿童炎症性肠病的治疗:微生物和饮食的联系
传统疗法
尽管在理解IBD的发病机制方面取得了上述进展,但目前大多数IBD的治疗方法主要是通过抑制免疫系统来减少炎症。pIBD的治疗在pCD和pUC之间有一些差异,但两者都可以使用糖皮质激素(glucocorticoids,GCS)、硫嘌呤、甲氨蝶呤、氨基水杨酸(aminosalicylates,5-ASA)和生物治疗 [200,201]。饮食疗法是目前针对pCD的推荐疗法,其独特之处是不抑制免疫系统,而全肠内营养(EEN)被认为是治疗腔内pCD的一线疗法(下文详细讨论)[201]。5-ASAs是抗炎药物,建议作为诱导和维持轻度或中度pUC缓解的一线治疗药物,而GCS是诱导pCD和pUC缓解的第二选择 [200,201]。GCS具有类固醇依赖的风险,并且有增加感染、糖尿病、骨质疏松和白内障风险等多种副作用 [202]。当5-ASA和硫嘌呤不能控制疾病时,推荐使用抗TNF等生物疗法诱导和维持缓解慢性活性或类固醇依赖/耐药pUC [200]。生物制剂是从另一种生物体或病毒中纯化出来的化合物 [203]。在pUC中,目前不建议使用FMT和抗生素; 但是像 VSL#3(几种乳酸杆菌、双歧杆菌和唾液链球菌嗜热亚种的混合物)或大肠杆菌 Nissle 1917 等益生菌被批准作为轻度pUC的辅助治疗,但效果有限。需要强有力的随机对照试验(randomized controlled trial,RCT) 证明益生菌是否比安慰剂更有效 [204]。
目前,有关治疗效果对宿主-微生物相互作用的影响仍然不清楚,即使在针对微生物群的治疗中也是如此。在正常和炎症条件下,GCS对动物的肠道微生物群落的改变作用包括增加双歧杆菌和乳酸杆菌种类,以及清除有粘蛋白降解功能的菌脱铁杆菌科 [204]。在pUC患者中,GCS的使用与持续缓解患者放线菌丰度的增加有关,并伴有梭状芽胞杆菌OTU的减少 [111]。硫嘌呤可能增加黏膜细菌数量和粘附,降低粪便细菌多样性和丰富度,并在体外抑制鸟分枝杆菌亚种副结核 [204],由于与分枝杆菌肠炎的病症相似,这也让一些人认为鸟分枝杆菌副结核在CD中发挥了致病作用 [205]。FMT最常用于复发性CDI,目前正在研究其在IBD中的效用,尽管其成功率低于CDI,但在UC和CD中显示出一些前景 [187]。与CDI相比,IBD的宿主-微生物关系的复杂性可能解释了迄今为止FMT和IBD缓解之间所观察到的混合结果和较弱的关系 [204]。如上所述,需要了解每个患者存在的独特微生物失调,从而对微生物组进行适当的治疗性修饰 (图1)。
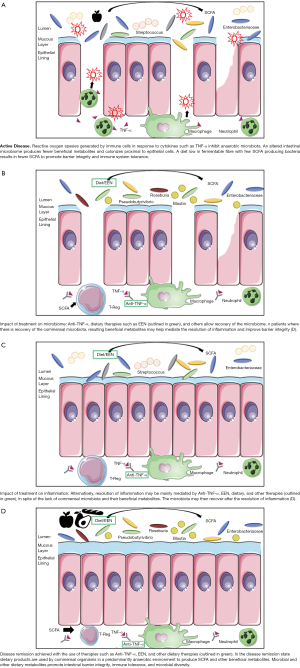
抗肿瘤坏死因子疗法
当患者进行了免疫调节治疗仍存在持续活动性管腔疾病、活动性类固醇难治性疾病、活动性肛周瘘管疾病,以及出现伴有生长迟缓的严重疾病,抗 TNF-α 治疗被推荐用于 pCD 患者 [201]。在pUC中,抗TNF-α治疗推荐用于不受5-ASA和硫嘌呤控制的慢性活动性或类固醇依赖性疾病 [200]。IFX和阿达木单抗(Adalimumab,ADA)是单克隆抗体(monoclonal antibodies,MAbs),可以识别膜结合和分泌型TNF-α [206]。膜结合TNF可能比分泌的可溶性TNF发挥更大的作用;优先阻断可溶性TNF的药物对IBD治疗尚未显示出效果 [193]。IFX阻断TNF的作用导致T细胞凋亡和促使免疫细胞流入肠道的细胞因子、趋化因子和粘附分子减少 [207]。然而,TNF-α和粘膜愈合之间的关系相当复杂,因为TNF-α敲除小鼠更有可能在DSS暴露下发生结肠炎 [99]。此外,益生菌VSL#3降低体外炎症回肠组织的肠通透性的能力在小鼠体内显示为TNF依赖性 [208]。无论如何,pIBD患者体内抗TNF治疗已被证明可以改善肠道上皮屏障并促进粘膜愈合 [209,210]。抗TNF-α的风险和副作用包括感染风险的增加,pCD的反常银屑病和输液反应 [16]。输液反应包括抗IFX或ADA抗体的产生,这与药物有效性的恶化有关 [211]。
pCD患者经过8周的抗TNF治疗后,患者的粪便微生物群与健康对照组更接近 [65]。与健康对照组相比,患有活动性疾病的pCD患者的相对属丰度降低了近三分之一,其中包括罗氏菌、瘤胃球菌、阿克曼氏菌、普氏菌、粪球菌和真杆菌 [65]。与基线相比,IFX治疗pCD后香农多样性指数和OTU数量增加,而和对照组的微生物类群差异更小 [212]。Wang等人发现,对IFX治疗有持续反应的pCD患者(这里定义为在随访期间PCDAI评分≤10)也显示IFX治疗后SCFA产生菌的属水平增加,包括布劳特氏菌、长栖粪杆菌、毛螺菌和罗氏菌 [158]。如果这些变化是治疗成功的原因或结果,那么抗 TNF-α 治疗中微生物群的一致变化还有待观察。由于IFX治疗是通过静脉注射,而ADA是通过皮下注射,因此与宿主肠道微生物组均没有直接的相互作用。由此产生的微生物变化可能是由于通过活性氧或抗菌肽等介质减少炎症而改变了宿主-微生物的相互作用,或者变化可能反映了伴随治疗的其他变化,例如饮食。
饮食疗法
EEN疗法是目前诱导pCD缓解的一线疗法,该疗法使用液体膳食替代饮料,并在6-8周内禁止摄入水以外其他食物和饮料 [213-215]。EEN最常用的配方包括Modulen IBD®,Ensure/Ensure Plus®,Pediasure®或Nutren/Nutren Junior®;配比公式可以是单质或混合 [216]。虽然这种治疗的安全性很好,且不担心副作用,但EEN对患者带来了相当大的困难。最常见的困难包括饮食的适口性和单调性,以及成本 [216]。EEN确切的作用机制虽然尚未阐明,但是理论解释包括免疫和/或微生物调节,去除许多膳食抗原和其他“有害食物”(如防腐剂和乳化剂),改善肠道屏障功能以及减少炎症等,其他人也对此进行了详细综述 [163,213,214]。
EEN疗法在开始后的一周内显著影响微生物群 [65],初步研究表明对EEN有反应和无反应的微生物群有不同的变化。当比较接受EEN疗法和皮质类固醇诱导疗法的新确诊的pUC和pCD患者时,发现缓解比疗法或疾病亚型更能预测患者微生物群的变化 [217]。Kaakoush等人发现对EEN有反应的患者的OTUs比无反应的患者下降更大(反应定义为治疗8至12周后PCDAI <10) [218]。Leach等人发现采用EEN治疗后拟杆菌和普雷沃氏菌种类减少最多的患者的PCDAI也表现出改善,甚至在EEN完成后4个月微生物群的变化仍持续存在 [219]。在Quince等人的一项研究中发现,EEN导致双歧杆菌、瘤胃球菌和粪钙杆菌减少;香农多样性指数在EEN中也有所下降,但在EEN完成两个月后恢复到治疗前水平 [220]。OTU和多样性的减少可能反映了这种流质饮食中缺乏纤维素 [219,211];基线时出现的与菌群失调相关类群数量的减少可能是治疗机制的重要组成部分或同质饮食的副作用。基线微生物对饮食的这种剧烈变化的反应可能是治疗反应的关键决定因素 [222]。
饮食疗法在pCD中的应用正在迅速扩大,其中一些正在对pUC进行试验,但许多与诱导缓解的成功疗法相关的微生物变化在有效的饮食疗法中是一致的。CD-TREAT是治疗pCD的另一种新的饮食疗法,它试图通过重建EEN 饮食排除,并与 EEN 相比诱导类似的微生物转变 [223]。克罗恩病排除饮食(Crohn’s Disease Exclusion diet, CDED)包括水果、蔬菜、碳水化合物和肉类等天然食物,类似于ENN可以诱导双歧杆菌和普雷沃氏菌减少,以及增加罗斯氏菌等微生物改变,后者通常在pIBD中减少且是一种已知的产丁酸菌 [65,158,224]。EEN和CDED在三周以内均可在80%的pCD病例中诱导快速反应 [225]。目前还不清楚这些微生物的变化是否在一定程度上决定了治疗的有效性,或者是导致了成功治疗和疾病缓解的结果。
扩大饮食疗法的范围
饮食干预仍然是pCD患者的安全治疗选择;但是,人们对饮食-宿主-微生物之间的许多相互作用仍然知之甚少。膳食纤维和蛋白质在这一关系中发挥着明显的作用,因为宿主微生物负责这些成分的发酵和分解,形成上面讨论的一些有益的产物 [62,226-228]。Williams等人对这些发酵产物的健康益处进行了深入的综述 [229]。研究表明典型的西式饮食导致IBD发病率显著上升 [2],其每天仅含有10-20克膳食纤维,而不是推荐的26-38克,这限制了许多肠道微生物的碳和能量来源 [230]。当我们仔细检查一组单独的膳食纤维时,则会增加更多的复杂性,这些纤维已发展成包括非淀粉多糖(如纤维素、果胶)、非消化性寡糖(如低聚果糖、半乳糖)、非碳水化合物聚合物(如多酚、木酚)、以及被认为是动物来源的碳水化合物(如几丁质)[231]。
膳食纤维通常根据其水溶性因子进行分类,其中可溶性纤维(果胶、阿拉伯木聚糖、β-葡聚糖、菊粉、低聚果糖、低聚半乳糖和木葡聚糖)比非可溶性纤维(纤维素和木质素)更容易作为微生物的营养来源 [61,229,232]。膳食纤维的微生物发酵涉及在协调群落中工作的各种物种,其中每种微生物在发酵途径中都发挥着关键作用,依靠协作生物来完成这一过程 [233-236]。许多患者在食用膳食纤维后症状恶化,如上所述菌群失调是IBD的明显特征;IBD患者纤维发酵有益产物的产生可能被改变或减少 [118,237-240]。这表明精确调整IBD患者的纤维摄入量以确保其独特微生物群的发酵能力,可能有助于提高有益纤维发酵产品的健康水平。尽管一些临床医生提倡在这些患者中减少纤维消耗或避免纤维摄入,但目前没有证据支持将其作为IBD的广泛治疗方法 [241,242]。当有能够发酵的微生物存在时,补充纤维可能对IBD产生有益的影响;IBD动物模型显示,与单纯使用益生元相比,给予益生菌/基于纤维的益生元可降低C-反应蛋白(C-reactive protein,CRP)水平,改善疾病活动性和组织学评分 [243]。对于 IBD 患者益生元的人体研究也显示了类似的结果 [244,245]。然而,目前我们对特定纤维类型在 IBD 中的确切作用的理解仍然知之甚少。
半纤维素阿拉伯木聚糖(arabinoxylan,AX)是饮食中纤维的重要组成部分,它主要来自黑麦、小麦、燕麦、大麦、大米、高粱等谷类和一些豆类 [246-251]。一些微生物将AX发酵成SCFA,如乙酸、丙酸、丁酸和乙醇 [249,252-254]。临床试验发现AX通过刺激盲肠粘蛋白层减少了炎症标志物,成人UC得到改善且无显著副作用,从而证明AX具有保护作用 [255-259]。乳酸杆菌、肠球菌和双歧杆菌可以发酵存在于蘑菇、燕麦和大麦等多种植物和真菌细胞壁中的β-葡聚糖纤维 [260-263]。β-葡聚糖摄入量对IBD组织学评分的影响高度依赖于纤维来源,而迄今为止最有效的纤维来源是杏鲍菇 [264]。同样,β-果聚糖通常存在于根茎、洋蓟、香蕉、小麦、洋葱和大蒜中,主要是由产生醋酸和醋酸转化丁酸的严格厌氧细菌发酵的,可以起到抗炎作用 [265-267]。纤维果胶作为抗炎治疗性膳食纤维较少受到关注,但存在于多种水果和蔬菜中,并且很容易被许多微生物发酵(拟杆菌、普氏菌、芽孢杆菌、农杆菌、假单胞菌、罗氏杆菌、菊欧文氏菌和酵母菌)[268-276]。果胶几乎完全发酵成各种SCFAs [268-276]。尽管动物模型已经证明了纤维素发酵生产SCFA的有益作用,但琥珀酸已被证实与促进炎症和细菌入侵有关,提示了患者描述的对食用膳食纤维敏感的可能机制 [140,141,278-280]。
每个 pIBD 患者的宿主微生物组和膳食纤维之间的独特相互作用可能有助于解释各种饮食疗法的一些机制,并突出未来发展和饮食调整的潜在途径。有关pIBD 目前和预期的饮食-宿主-微生物相互作用的理解可以更好地定制和开发改良疗法。建立在这些原则基础上的新方法将允许在考虑治疗选择时纳入患者的基线微生物组/代谢组状态。例如,如果减少特定的SCFAs,可以添加负责生产它们的微生物,或者添加富含益生元的饮食,从而促进这些微生物的生长。
结论和未来展望
随着pIBD全球患病率的增加,有必要更好地了解这些疾病的发病机制,以防止这些患者出现不良预后。相关的遗传学、免疫系统变化、饮食和微生物组表明,宿主微生物关系的改变对于疾病的发展和持续很重要。目前以及新的治疗方法显著地改变了这种关系,在某些情况下改善这种状况可能是实现病情缓解的必要条件。由于患者微生物组的代谢能力可能会影响其对治疗的反应,因此对患者的宿主-微生物基线关系的评估为治疗开发和饮食调整提供了一个新的领域。随着正在进行的研究阐明了这种饮食-宿主-微生物的关系,考虑到基线微生物组状态,通过饮食和其他疗法针对微生物群成分及其能力的改变可能会提升儿童IBD 患者的疾病缓解率和改善治疗结果。
Acknowledgments
Funding: SD was supported by student awards from Canadian Institutes of Health Research (CIHR); the Women & Children’s Health Research Institute; and the University of Alberta. HA was supported by a CIHR Fellowship. The Wine lab was funded by CIHR and the Weston Family Foundation.
Footnote
Reporting Checklist: The authors have completed the narrative review checklist. Available at http://dx.doi.org/10.21037/dmr-20-160
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/dmr-20-160). Dr. Wine reports personal fees from AbbVie, Janssen, Nestle Health Sciences and Mead Johnson Nutrition, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2017;390:2769-78. [Crossref] [PubMed]
- Coward S, Clement F, Benchimol EI, et al. Past and Future Burden of Inflammatory Bowel Diseases Based on Modeling of Population-Based Data. Gastroenterology 2019;156:1345-53.e4. [Crossref] [PubMed]
- Roberts SE, Thorne K, Thapar N, et al. A systematic review and meta-analysis of paediatric inflammatory bowel disease incidence and prevalence across Europe. J Crohns Colitis 2020;14:1119-48. [Crossref] [PubMed]
- Kelsen J, Baldassano RN. Inflammatory bowel disease: the difference between children and adults. Inflamm Bowel Dis 2008;14:S9-S11. [Crossref] [PubMed]
- Ye Y, Manne S, Treem WR, et al. Prevalence of inflammatory bowel disease in pediatric and adult populations: recent estimates from large national databases in the United States, 2007–2016. Inflamm Bowel Dis 2020;26:619-25. [PubMed]
- Shaw KA, Cutler DJ, Okou D, et al. Genetic variants and pathways implicated in a pediatric inflammatory bowel disease cohort. Genes Immun 2019;20:131-42. [Crossref] [PubMed]
- Salvador-Martín S, Bossacoma F, Pujol-Muncunill G, et al. Genetic Predictors of Long-term Response to Antitumor Necrosis Factor Agents in Pediatric Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr 2020;71:508-15. [Crossref] [PubMed]
- Lin Z, Wang Z, Hegarty JP, et al. Genetic association and epistatic interaction of the interleukin-10 signaling pathway in pediatric inflammatory bowel disease. World J Gastroenterol 2017;23:4897. [Crossref] [PubMed]
- Penagini F, Dilillo D, Borsani B, et al. Nutrition in pediatric inflammatory bowel disease: from etiology to treatment. A systematic review. Nutrients 2016;8:334. [Crossref] [PubMed]
- Strisciuglio C, Giugliano F, Martinelli M, et al. Impact of environmental and familial factors in a cohort of pediatric patients with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2017;64:569-74. [Crossref] [PubMed]
- Vitale A, Strisciuglio C, Vitale S, et al. Increased frequency of regulatory T cells in pediatric inflammatory bowel disease at diagnosis: a compensative role? Pediatr Res 2020;87:853-61. [Crossref] [PubMed]
- Aujnarain A, Mack DR, Benchimol EI. The role of the environment in the development of pediatric inflammatory bowel disease. Curr Gastroenterol Rep 2013;15:326. [Crossref] [PubMed]
- Kolho K-L, Korpela K, Jaakkola T, et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol 2015;110:921-30. [Crossref] [PubMed]
- Wright EK, Kamm MA, Teo SM, et al. Recent advances in characterizing the gastrointestinal microbiome in Crohn's disease: a systematic review. Inflamm Bowel Dis 2015;21:1219-28. [PubMed]
- Mamula P, Telega GW, Markowitz JE, et al. Inflammatory bowel disease in children 5 years of age and younger. Am J Gastroenterol 2002;97:2005-10. [Crossref] [PubMed]
- Corica D, Romano C. Biological therapy in pediatric inflammatory bowel disease. J Clin Gastroenterol 2017;51:100-10. [Crossref] [PubMed]
- Wong K, Isaac DM, Wine E. Growth Delay in Inflammatory Bowel Diseases: Significance, Causes, and Management. Dig Dis Sci 2021;66:954-64. [Crossref] [PubMed]
- MacKner LM, Crandall WV, Szigethy EM. Psychosocial functioning in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2006;12:239-44. [Crossref] [PubMed]
- Polito JM, Childs B, Mellits ED, et al. Crohn's disease: Influence of age at diagnosis on site and clinical type of disease. Gastroenterology 1996;111:580-6. [Crossref] [PubMed]
- Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 2011;17:1314-21. [Crossref] [PubMed]
- Vernier–Massouille G, Balde M, Salleron J, et al. Natural history of pediatric Crohn's disease: a population-based cohort study. Gastroenterology 2008;135:1106-13. [Crossref] [PubMed]
- Guariso G, Gasparetto M, Dalla Pozza LV, et al. Inflammatory bowel disease developing in paediatric and adult age. J Pediatr Gastroenterol Nutr 2010;51:698-707. [Crossref] [PubMed]
- Kim BJ, Song SM, Kim KM, et al. Characteristics and trends in the incidence of inflammatory bowel disease in Korean children: a single-center experience. Dig Dis Sci 2010;55:1989-95. [Crossref] [PubMed]
- Sawczenko A, Sandhu BK. Presenting features of inflammatory bowel disease in Great Britain and Ireland. Arch Dis Child 2003;88:995-1000. [Crossref] [PubMed]
- Martins AL, Volpato RA, da Penha Zago-Gomes M. The prevalence and phenotype in Brazilian patients with inflammatory bowel disease. BMC Gastroenterol 2018;18:87. [Crossref] [PubMed]
- Zeng Z, Zhu Z, Yang Y, et al. Incidence and clinical characteristics of inflammatory bowel disease in a developed region of Guangdong Province, China: A prospective population-based study. J Gastroenterol Hepatol 2013;28:1148-53. [Crossref] [PubMed]
- Frolkis AD, Dykeman J, Negrón ME, et al. Risk of surgery for inflammatory bowel diseases has decreased over time: a systematic review and meta-analysis of population-based studies. Gastroenterology 2013;145:996-1006. [Crossref] [PubMed]
- Ross SC, Strachan J, Russell RK, et al. Psychosocial functioning and health-related quality of life in paediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2011;53:480-8. [Crossref] [PubMed]
- Malham M, Jakobsen C, Paerregaard A, et al. The incidence of cancer and mortality in paediatric onset inflammatory bowel disease in Denmark and Finland during a 23-year period: a population-based study. Aliment Pharmacol Ther 2019;50:33-9. [Crossref] [PubMed]
- Kugathasan S, Judd RH, Hoffmann RG, et al. Epidemiologic and clinical characteristics of children with newly diagnosed inflammatory bowel disease in Wisconsin: a statewide population-based study. J Pediatr 2003;143:525-31. [Crossref] [PubMed]
- Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527-39. [Crossref] [PubMed]
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307-17. [Crossref] [PubMed]
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut 2013;62:1505-10. [Crossref] [PubMed]
- Cleynen I, González JR, Figueroa C, et al. Genetic factors conferring an increased susceptibility to develop Crohn's disease also influence disease phenotype: results from the IBDchip European Project. Gut 2013;62:1556-65. [Crossref] [PubMed]
- Al Nabhani Z, Dietrich G, Hugot JP, et al. Nod2: The intestinal gate keeper. PLoS Pathog 2017;13:e1006177 [Crossref] [PubMed]
- Bonen DK, Ogura Y, Nicolae DL, et al. Crohn's disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology 2003;124:140-6. [Crossref] [PubMed]
- Tan G, Zeng B, Zhi F-C. Regulation of human enteric α-defensins by NOD2 in the Paneth cell lineage. Eur J Cell Biol 2015;94:60-6. [Crossref] [PubMed]
- Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001;411:603-6. [Crossref] [PubMed]
- Hisamatsu T, Suzuki M, Reinecker H-C, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 2003;124:993-1000. [Crossref] [PubMed]
- Schäffler H, Geiss D, Gittel N, et al. Mutations in the NOD2 gene are associated with a specific phenotype and lower anti-tumor necrosis factor trough levels in Crohn's disease. J Dig Dis 2018;19:678-84. [Crossref] [PubMed]
- Wine E, Reif SS, Leshinsky-Silver E, et al. Pediatric Crohn's disease and growth retardation: the role of genotype, phenotype, and disease severity. Pediatrics 2004;114:1281-6. [Crossref] [PubMed]
- Leshchiner ES, Rush JS, Durney MA, et al. Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease. Proc Natl Acad Sci U S A 2017;114:11392-7. [Crossref] [PubMed]
- Hsu Y-MS, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol 2007;8:198-205. [Crossref] [PubMed]
- Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 2019;25:377-88.e6. [Crossref] [PubMed]
- Prager M, Büttner J, Haas V, et al. The JAK2 variant rs10758669 in Crohn’s disease: altering the intestinal barrier as one mechanism of action. Int J Colorectal Dis 2012;27:565-73. [Crossref] [PubMed]
- Hedl M, Proctor DD, Abraham C. JAK2 disease-risk variants are gain of function and JAK signaling threshold determines innate receptor-induced proinflammatory cytokine secretion in macrophages. J Immunol 2016;197:3695-704. [Crossref] [PubMed]
- Ghoreschi K, Jesson MI, Li X, et al. Modulation of innate and adaptive immune responses by tofacitinib (CP-690,550). J Immunol 2011;186:4234-43. [Crossref] [PubMed]
- Feagan B. Update on tofacitinib for inflammatory bowel disease. Gastroenterol Hepatol 2016;12:572. [PubMed]
- Vancamelbeke M, Vanuytsel T, Farré R, et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm Bowel Dis 2017;23:1718-29. [Crossref] [PubMed]
- Darsigny M, Babeu JP, Dupuis AA, et al. Loss of hepatocyte-nuclear-factor-4α affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One 2009;4:e7609 [Crossref] [PubMed]
- Mohanan V, Nakata T, Desch AN, et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 2018;359:1161-6. [Crossref] [PubMed]
- Manzanillo P, Mouchess M, Ota N, et al. Inflammatory bowel disease susceptibility gene C1ORF106 regulates intestinal epithelial permeability. ImmunoHorizons 2018;2:164-71. [Crossref] [PubMed]
- Fujimoto K, Kinoshita M, Tanaka H, et al. Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol 2017;10:446-59. [Crossref] [PubMed]
- Chang J, Leong RW, Wasinger VC, et al. Impaired intestinal permeability contributes to ongoing bowel symptoms in patients with inflammatory bowel disease and mucosal healing. Gastroenterology 2017;153:723-31.e1. [Crossref] [PubMed]
- Kiesslich R, Duckworth C, Moussata D, et al. Local barrier dysfunction identified by confocal laser endomicroscopy predicts relapse in inflammatory bowel disease. Gut 2012;61:1146-53. [Crossref] [PubMed]
- Martini E, Krug SM, Siegmund B, et al. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol 2017;4:33-46. [Crossref] [PubMed]
- Peters LA, Perrigoue J, Mortha A, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49:1437. [Crossref] [PubMed]
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016;14:e1002533 [Crossref] [PubMed]
- Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol 2015;31:69. [Crossref] [PubMed]
- Kamada N, Núñez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 2014;146:1477-88. [Crossref] [PubMed]
- Armstrong H, Mander I, Zhang Z, et al. Not all fibres are born equal; variable response to dietary fibre subtypes in IBD. Front Pediatr 2021;8:620189 [Crossref] [PubMed]
- Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014;146:1449-58. [Crossref] [PubMed]
- Chehoud C, Albenberg LG, Judge C, et al. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis 2015;21:1948-56. [Crossref] [PubMed]
- Mukhopadhya I, Hansen R, Meharg C, et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect 2015;17:304-10. [Crossref] [PubMed]
- Lewis James D, Chen Eric Z, Baldassano Robert N, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015;18:489-500. [Crossref] [PubMed]
- Imai T, Inoue R, Kawada Y, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol 2019;54:149-59. [Crossref] [PubMed]
- Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039-48. [Crossref] [PubMed]
- Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 2008;43:831-41. [Crossref] [PubMed]
- Birimberg-Schwartz L, Wilson DC, Kolho KL, et al. pANCA and ASCA in children with IBD-unclassified, Crohn's colitis, and ulcerative colitis—a longitudinal report from the IBD Porto Group of ESPGHAN. Inflamm Bowel Dis 2016;22:1908-14. [Crossref] [PubMed]
- Lam S, Zuo T, Ho M, et al. fungal alterations in inflammatory bowel diseases. Aliment Pharmacol Ther 2019;50:1159-71. [Crossref] [PubMed]
- Liang G, Conrad MA, Kelsen JR, et al. Dynamics of the Stool Virome in Very Early-Onset Inflammatory Bowel Disease. J Crohns Colitis 2020;14:1600-10. [Crossref] [PubMed]
- Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447-60. [Crossref] [PubMed]
- Bolsega S, Basic M, Smoczek A, et al. Composition of the intestinal microbiota determines the outcome of virus-triggered colitis in mice. Front Immunol 2019;10:1708. [Crossref] [PubMed]
- Wang W, Jovel J, Halloran B, et al. Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm Bowel Dis 2015;21:1419-27. [Crossref] [PubMed]
- Babickova J, Gardlik R. Pathological and therapeutic interactions between bacteriophages, microbes and the host in inflammatory bowel disease. World J Gastroenterol: WJG 2015;21:11321. [Crossref] [PubMed]
- Clooney AG, Sutton TD, Shkoporov AN, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019;26:764-78.e5. [Crossref] [PubMed]
- Ungaro F, Massimino L, D’Alessio S, et al. The gut virome in inflammatory bowel disease pathogenesis: From metagenomics to novel therapeutic approaches. United European Gastroenterol J 2019;7:999-1007. [Crossref] [PubMed]
- Lopetuso LR, Ianiro G, Scaldaferri F, et al. Gut virome and inflammatory bowel disease. Inflamm Bowel Dis 2016;22:1708-12. [Crossref] [PubMed]
- Prosberg M, Bendtsen F, Vind I, et al. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol 2016;51:1407-15. [Crossref] [PubMed]
- Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010;139:1844-54.e1. [Crossref] [PubMed]
- Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosaassociated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis 2009;15:653-60. [Crossref] [PubMed]
- Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2008;2:716-27. [Crossref] [PubMed]
- Ng SC, Woodrow S, Patel N, et al. Role of genetic and environmental factors in British twins with inflammatory bowel disease. Inflamm Bowel Dis 2012;18:725-36. [Crossref] [PubMed]
- Virta L, Auvinen A, Helenius H, et al. Association of repeated exposure to antibiotics with the development of pediatric Crohn’s disease—a nationwide, register-based Finnish case-control study. Am J Epidemiol 2012;175:775-84. [Crossref] [PubMed]
- Theochari NA, Stefanopoulos A, Mylonas KS, et al. Antibiotics exposure and risk of inflammatory bowel disease: a systematic review. Scand J Gastroenterol 2018;53:1-7. [Crossref] [PubMed]
- Zuo T, Zhang F, Lui GC, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020;159:944-55.e8. [Crossref] [PubMed]
- Levin AM, Sitarik AR, Havstad SL, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep 2016;6:31775. [Crossref] [PubMed]
- Korpela K, Salonen A, Virta LJ, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016;7:10410. [Crossref] [PubMed]
- Nermes M, Endo A, Aarnio J, et al. Furry pets modulate gut microbiota composition in infants at risk for allergic disease. J Allergy Clin Immunol 2015;136:1688. [Crossref] [PubMed]
- Tun HM, Konya T, Takaro TK, et al. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 2017;5:40. [Crossref] [PubMed]
- Lu K, Cable PH, Abo RP, et al. Gut microbiome perturbations induced by bacterial infection affect arsenic biotransformation. Chem Res Toxicol 2013;26:1893-903. [Crossref] [PubMed]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559-63. [Crossref] [PubMed]
- Johnson AJ, Vangay P, Al-Ghalith GA, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 2019;25:789-802.e5. [Crossref] [PubMed]
- Alam MT, Amos GC, Murphy AR, et al. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog 2020;12:1-8. [Crossref] [PubMed]
- Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382-92. [Crossref] [PubMed]
- Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res 2017;10:63. [Crossref] [PubMed]
- Zhou Y, Zhi F. Lower level of bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-analysis. Biomed Res Int 2016;2016:5828959 [Crossref] [PubMed]
- Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731-6. [Crossref] [PubMed]
- Sánchez de Medina F, Romero-Calvo I, Mascaraque C, et al. Intestinal Inflammation and Mucosal Barrier Function. Inflamm Bowel Dis 2014;20:2394-404. [Crossref] [PubMed]
- Quévrain E, Maubert M, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016;65:415-25. [Crossref] [PubMed]
- Arnal M-E, Lalles J-P. Gut epithelial inducible heat-shock proteins and their modulation by diet and the microbiota. Nutr Rev 2016;74:181-97. [Crossref] [PubMed]
- Xu J, Liang R, Zhang W, et al. Faecalibacterium prausnitzii-derived microbial anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J Diabetes 2020;12:224-36. [Crossref] [PubMed]
- Lallès J-P. Microbiota-host interplay at the gut epithelial level, health and nutrition. J Anim Sci Biotechnol 2016;7:66. [Crossref] [PubMed]
- Hedin CR, van der Gast CJ, Stagg AJ, et al. The gut microbiota of siblings offers insights into microbial pathogenesis of inflammatory bowel disease. Gut Microbes 2017;8:359-65. [Crossref] [PubMed]
- Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011;141:227-36. [Crossref] [PubMed]
- Brand EC, Klaassen MA, Gacesa R, et al. Healthy cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterology 2021; Epub ahead of print. [Crossref] [PubMed]
- Nishino K, Nishida A, Inoue R, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol 2018;53:95-106. [Crossref] [PubMed]
- Shah R, Cope JL, Nagy-Szakal D, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes 2016;7:384-96. [Crossref] [PubMed]
- Fritsch J, Abreu MT. Candida in IBD: Friend or Foe? Cell Host Microbe 2020;27:689-91. [Crossref] [PubMed]
- Michail S, Durbin M, Turner D, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis 2012;18:1799-808. [Crossref] [PubMed]
- Schirmer M, Denson L, Vlamakis H, et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 2018;24:600-10.e4. [Crossref] [PubMed]
- Ryan FJ, Ahern A, Fitzgerald R, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun 2020;11:1512. [Crossref] [PubMed]
- Alipour M, Zaidi D, Valcheva R, et al. Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J Crohns Colitis 2016;10:462-71. [Crossref] [PubMed]
- Roy U, Gálvez EJ, Iljazovic A, et al. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep 2017;21:994-1008. [Crossref] [PubMed]
- Rengarajan S, Vivio EE, Parkes M, et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes 2020;11:405-20. [Crossref] [PubMed]
- Coufal S, Galanova N, Bajer L, et al. Inflammatory bowel disease types differ in markers of inflammation, gut barrier and in specific anti-bacterial response. Cells 2019;8:719. [Crossref] [PubMed]
- Palm NW, De Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000-10. [Crossref] [PubMed]
- Armstrong H, Alipour M, Valcheva R, et al. Host immunoglobulin G selectively identifies pathobionts in pediatric inflammatory bowel diseases. Microbiome 2019;7:1-17. [Crossref] [PubMed]
- Moor K, Diard M, Sellin ME, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017;544:498-502. [Crossref] [PubMed]
- Wilmore JR, Gaudette BT, Atria DG, et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe 2018;23:302-11.e3. [Crossref] [PubMed]
- Castro-Dopico T, Dennison TW, Ferdinand JR, et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 2019;50:1099-114.e10. [Crossref] [PubMed]
- Adams RJ, Heazlewood SP, Gilshenan KS, et al. IgG antibodies against common gut bacteria are more diagnostic for Crohn's disease than IgG against mannan or flagellin. Am J Gastroenterol 2008;103:386-96. [Crossref] [PubMed]
- Lin R, Chen H, Shu W, et al. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med 2018;16:359. [Crossref] [PubMed]
- Torres J, Petralia F, Sato T, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology 2020;159:96-104. [Crossref] [PubMed]
- Zaidi D, Bording-Jorgensen M, Huynh HQ, et al. Increased Epithelial Gap Density in the Noninflamed Duodenum of Children With Inflammatory Bowel Diseases. J Pediatr Gastroenterol Nutr 2016;63:644-50. [Crossref] [PubMed]
- Vivinus-Nebot M, Frin-Mathy G, Bzioueche H, et al. Functional bowel symptoms in quiescent inflammatory bowel diseases: role of epithelial barrier disruption and low-grade inflammation. Gut 2014;63:744-52. [Crossref] [PubMed]
- Hollander D, Vadheim CM, Brettholz E, et al. Increased intestinal permeability in patients with Crohn's disease and their relatives: a possible etiologic factor. Ann Intern Med 1986;105:883-5. [Crossref] [PubMed]
- Munkholm P, Langholz E, Hollander D, et al. Intestinal permeability in patients with Crohn's disease and ulcerative colitis and their first degree relatives. Gut 1994;35:68-72. [Crossref] [PubMed]
- Olson TS, Reuter BK, Scott KG, et al. The primary defect in experimental ileitis originates from a nonhematopoietic source. J Exp Med 2006;203:541-52. [Crossref] [PubMed]
- Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol 2013;14:660. [Crossref] [PubMed]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 2007;19:59-69. [Crossref] [PubMed]
- Armstrong H, Bording-Jorgensen M, Dijk S, et al. The complex interplay between chronic inflammation, the microbiome, and cancer: understanding disease progression and what we can do to prevent it. Cancers 2018;10:83. [Crossref] [PubMed]
- Haverson K, Rehakova Z, Sinkora J, et al. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol 2007;119:243-53. [Crossref] [PubMed]
- Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci 2010;15:25. [Crossref] [PubMed]
- Kozakova H, Schwarzer M, Tuckova L, et al. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell Mol Immunol 2016;13:251-62. [Crossref] [PubMed]
- Schicho R, Shaykhutdinov R, Ngo J, et al. Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J Proteome Res 2012;11:3344-57. [Crossref] [PubMed]
- Stephens NS, Siffledeen J, Su X, et al. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J Crohns Colitis 2013;7:e42-8. [Crossref] [PubMed]
- Kolho KL, Pessia A, Jaakkola T, et al. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis 2017;11:321-34. [PubMed]
- Martin F-P, Ezri J, Cominetti O, et al. Urinary metabolic phenotyping reveals differences in the metabolic status of healthy and inflammatory bowel disease (IBD) children in relation to growth and disease activity. Int J Mol Med Sci 2016;17:1310. [Crossref] [PubMed]
- Zaidi D, Huynh HQ, Carroll MW, et al. Gut Microenvironment and Bacterial Invasion in Paediatric Inflammatory Bowel Diseases. J Pediatr Gastroenterol Nutr 2020;71:624-32. [Crossref] [PubMed]
- Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223-37. [Crossref] [PubMed]
- Martin FP, Su MM, Xie GX, et al. Urinary metabolic insights into host-gut microbial interactions in healthy and IBD children. World J Gastroenterol 2017;23:3643-54. [Crossref] [PubMed]
- Lees HJ, Swann JR, Wilson ID, et al. Hippurate: the natural history of a mammalian–microbial cometabolite. J Proteome Res 2013;12:1527-46. [Crossref] [PubMed]
- Sarosiek I, Schicho R, Blandon P, et al. Urinary metabolites as noninvasive biomarkers of gastrointestinal diseases: A clinical review. World J Gastrointest Oncol 2016;8:459. [Crossref] [PubMed]
- Pallister T, Jackson MA, Martin TC, et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci Rep 2017;7:13670. [Crossref] [PubMed]
- Williams HR, Cox JI, Walker DG, et al. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol 2009;104:1435-44. [Crossref] [PubMed]
- Williams HR, Cox IJ, Walker DG, et al. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn's disease. BMC Gastroenterol 2010;10:108. [Crossref] [PubMed]
- Huda-Faujan N, Abdulamir AS, Fatimah AB, et al. The impact of the level of the intestinal short chain Fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J 2010;4:53-8. [Crossref] [PubMed]
- Treem WR, Ahsan N, Shoup M, et al. Fecal short-chain fatty acids in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 1994;18:159-64. [Crossref] [PubMed]
- Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr 2008;100:297-305. [Crossref] [PubMed]
- Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569-73. [Crossref] [PubMed]
- Venegas DP, Marjorie K, Landskron G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol 2019;10:277. [Crossref] [PubMed]
- Ahmad M, Krishnan S, Ramakrishna B, et al. Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut 2000;46:493-9. [Crossref] [PubMed]
- Agus A, Denizot J, Thevenot J, et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci Rep 2016;6:19032. [Crossref] [PubMed]
- Ferrer-Picón E, Dotti I, Corraliza AM, et al. Intestinal inflammation modulates the epithelial response to butyrate in patients with inflammatory bowel disease. Inflamm Bowel Dis 2020;26:43-55. [Crossref] [PubMed]
- Cavaglieri CR, Nishiyama A, Fernandes LC, et al. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci 2003;73:1683-90. [Crossref] [PubMed]
- Segain JP, Raingeard de la Blétière D, Bourreille A, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn's disease. Gut 2000;47:397-403. [Crossref] [PubMed]
- Wang Y, Gao X, Ghozlane A, et al. Characteristics of Faecal Microbiota in Paediatric Crohn's Disease and Their Dynamic Changes During Infliximab Therapy. J Crohns Colitis 2018;12:337-46. [Crossref] [PubMed]
- Roediger W. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology 1982;83:424-9. [Crossref] [PubMed]
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 2001;81:1031-64. [Crossref] [PubMed]
- Pang T, Leach ST, Katz T, et al. Fecal biomarkers of intestinal health and disease in children. Front Pediatr 2014;2:6. [Crossref] [PubMed]
- Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol 2011;106:563-73. [Crossref] [PubMed]
- Levine A, Boneh RS, Wine E. Evolving role of diet in the pathogenesis and treatment of inflammatory bowel diseases. Gut 2018;67:1726-38. [Crossref] [PubMed]
- Niewiadomski O, Studd C, Wilson J, et al. Influence of food and lifestyle on the risk of developing inflammatory bowel disease. Intern Med J 2016;46:669-76. [Crossref] [PubMed]
- Rashvand S, Somi MH, Rashidkhani B, et al. Dietary protein intakes and risk of ulcerative colitis. Med J Islam Repub Iran 2015;29:253. [PubMed]
- Jantchou P, Morois S, Clavel-Chapelon F, et al. Animal protein intake and risk of inflammatory bowel disease: The E3N prospective study. Am J Gastroenterol 2010;105:2195-201. [Crossref] [PubMed]
- Khalili H, de Silva PS, Ananthakrishnan AN, et al. Dietary iron and heme iron consumption, genetic susceptibility, and risk of Crohn's disease and ulcerative colitis. Inflamm Bowel Dis 2017;23:1088-95. [Crossref] [PubMed]
- D'Souza S, Levy E, Mack D, et al. Dietary patterns and risk for Crohn's disease in children. Inflamm Bowel Dis 2008;14:367-73. [Crossref] [PubMed]
- Racine A, Carbonnel F, Chan SS, et al. Dietary patterns and risk of inflammatory bowel disease in Europe: results from the EPIC study. Inflamm Bowel Dis 2016;22:345-54. [Crossref] [PubMed]
- Lo C-H, Lochhead P, Khalili H, et al. Dietary inflammatory potential and risk of Crohn’s disease and ulcerative colitis. Gastroenterology 2020;159:873-83.e1. [Crossref] [PubMed]
- Howe A, Ringus DL, Williams RJ, et al. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. The ISME Journal 2016;10:1217-27. [Crossref] [PubMed]
- Singh RK, Chang H-W, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med 2017;15:73. [Crossref] [PubMed]
- Martinez-Medina M, Denizot J, Dreux N, et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 2014;63:116-24. [Crossref] [PubMed]
- Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: an update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol 2014;5:213. [Crossref] [PubMed]
- Goyal A, Yeh A, Bush BR, et al. Safety, clinical response, and microbiome findings following fecal microbiota transplant in children with inflammatory bowel disease. Inflamm Bowel Dis 2018;24:410-21. [Crossref] [PubMed]
- Caldeira LdF. Fecal microbiota transplantation in inflammatory bowel disease patients: A systematic review and meta-analysis. PLoS One 2020;15:e0238910 [Crossref] [PubMed]
- Bibbò S, Settanni CR, Porcari S, et al. Fecal microbiota transplantation: screening and selection to choose the optimal donor. J Clin Med 2020;9:1757. [Crossref] [PubMed]
- Fang H, Fu L, Wang J. Protocol for fecal microbiota transplantation in inflammatory bowel disease: a systematic review and meta-analysis. Biomed Res Int 2018;2018:8941340 [Crossref] [PubMed]
- Imdad A, Nicholson MR, Tanner-Smith EE, et al. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst Rev 2018;11:CD012774 [Crossref] [PubMed]
- Keshteli A, Millan B, Madsen K. Pretreatment with antibiotics may enhance the efficacy of fecal microbiota transplantation in ulcerative colitis: a meta-analysis. Mucosal Immunol 2017;10:565-6. [Crossref] [PubMed]
- Leonardi I, Paramsothy S, Doron I, et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe 2020;27:823-9.e3. [Crossref] [PubMed]
- Qazi T, Amaratunga T, Barnes EL, et al. The risk of inflammatory bowel disease flares after fecal microbiota transplantation: Systematic review and meta-analysis. Gut Microbes 2017;8:574-88. [Crossref] [PubMed]
- Khanna S, Vazquez-Baeza Y, González A, et al. Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 2017;5:55. [Crossref] [PubMed]
- Hourigan S, Chen L, Grigoryan Z, et al. Microbiome changes associated with sustained eradication of Clostridium difficile after single faecal microbiota transplantation in children with and without inflammatory bowel disease. Aliment Pharmacol Ther 2015;42:741-52. [Crossref] [PubMed]
- Gutin L, Piceno Y, Fadrosh D, et al. Fecal microbiota transplant for Crohn disease: A study evaluating safety, efficacy, and microbiome profile. United European Gastroenterol J 2019;7:807-14. [Crossref] [PubMed]
- Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017;8:238-52. [Crossref] [PubMed]
- Sunkara T, Rawla P, Ofosu A, et al. Fecal microbiota transplant–a new frontier in inflammatory bowel disease. J Inflamm Res 2018;11:321. [Crossref] [PubMed]
- D'Odorico I, Di Bella S, Monticelli J, et al. Role of fecal microbiota transplantation in inflammatory bowel disease. J Dig Dis 2018;19:322-34. [Crossref] [PubMed]
- Allegretti J, Eysenbach LM, El-Nachef N, et al. The current landscape and lessons from fecal microbiota transplantation for inflammatory bowel disease: past, present, and future. Inflamm Bowel Dis 2017;23:1710-7. [Crossref] [PubMed]
- Maxwell EC, Dawany N, Baldassano RN, et al. Diverting ileostomy for the treatment of severe, refractory, pediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2017;65:299-305. [Crossref] [PubMed]
- Keubler LM, Buettner M, Häger C, et al. A multihit model: colitis lessons from the interleukin-10–deficient mouse. Inflamm Bowel Dis 2015;21:1967-75. [Crossref] [PubMed]
- Schaubeck M, Clavel T, Calasan J, et al. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut 2016;65:225-37. [Crossref] [PubMed]
- Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol 2014;14:329-42. [Crossref] [PubMed]
- Rahman MM, McFadden G. Modulation of NF-κB signalling by microbial pathogens. Nat Rev Microbiol 2011;9:291-306. [Crossref] [PubMed]
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011;9:356-68. [Crossref] [PubMed]
- Comito D, Cascio A, Romano C. Microbiota biodiversity in inflammatory bowel disease. Ital J Pediatr 2014;40:32. [Crossref] [PubMed]
- Choung R, Princen F, Stockfisch T, et al. Serologic microbial associated markers can predict Crohn's disease behaviour years before disease diagnosis. Aliment Pharmacol Ther 2016;43:1300-10. [Crossref] [PubMed]
- Dubinsky MC, Kugathasan S, Mei L, et al. Increased immune reactivity predicts aggressive complicating Crohn's disease in children. Clin Gastroenterol Hepatol 2008;6:1105-11. [Crossref] [PubMed]
- Spencer EA, Davis SM, Mack DR, et al. Serologic reactivity reflects clinical expression of ulcerative colitis in children. Inflamm Bowel Dis 2018;24:1335-43. [Crossref] [PubMed]
- Turner D, Ruemmele FM, Orlanski-Meyer E, et al. Management of paediatric ulcerative colitis, part 1: ambulatory care—an evidence-based guideline from European Crohn's and Colitis Organization and European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2018;67:257-91. [Crossref] [PubMed]
- van Rheenen PF, Aloi M, Assa A, et al. The medical management of paediatric Crohn’s disease: an ECCO-ESPGHAN guideline update. J Crohns Colitis. 2020 Oct 7:jjaa161. [Epub ahead of print]. doi:
10.1093/ecco-jcc/jjaa161 . - Saag K, Furst D. Major side effects of systemic glucocorticoids. In: Matteson E, editor. UpToDate. UpToDate, Waltham, MA. (Accessed on May 24, 2020)2020.
- Agency EM. Remicade. 2020. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/remicade. Accessed June 15, 2020 2020.
- Nishida A, Inoue R, Inatomi O, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 2018;11:1-10. [Crossref] [PubMed]
- Naser SA, Ghobrial G, Romero C, et al. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 2004;364:1039-44. [Crossref] [PubMed]
- Danese S. Mechanisms of action of infliximab in inflammatory bowel disease: an anti-inflammatory multitasker. Dig Liver Dis 2008;40:S225-8. [Crossref] [PubMed]
- Pasparakis M. Regulation of tissue homeostasis by NF-κB signalling: implications for inflammatory diseases. Nat Rev Immunol 2009;9:778-88. [Crossref] [PubMed]
- Corridoni D, Pastorelli L, Mattioli B, et al. Probiotic bacteria regulate intestinal epithelial permeability in experimental ileitis by a TNF-dependent mechanism. PLoS One 2012;7:e42067 [Crossref] [PubMed]
- Nobile S, Gionchetti P, Rizzello F, et al. Mucosal healing in pediatric Crohn’s disease after anti-TNF therapy: a long-term experience at a single center. Eur J Gastroenterol Hepatol 2014;26:458-65. [Crossref] [PubMed]
- Hyams J, Damaraju L, Blank M, et al. Induction and maintenance therapy with infliximab for children with moderate to severe ulcerative colitis. Clin Gastroenterol Hepatol 2012;10:391-9.e1. [Crossref] [PubMed]
- Ungar B, Glidai Y, Yavzori M, et al. Association between infliximab drug and antibody levels and therapy outcome in pediatric inflammatory bowel diseases. J Pediatr Gastroenterol Nutr 2018;67:507-12. [Crossref] [PubMed]
- Kowalska-Duplaga K, Kapusta P, Gosiewski T, et al. Changes in the Intestinal Microbiota Are Seen Following Treatment with Infliximab in Children with Crohn’s Disease. J Clin Med 2020;9:687. [Crossref] [PubMed]
- Levine A, Wine E. Effects of enteral nutrition on Crohn’s disease: clues to the impact of diet on disease pathogenesis. Inflamm Bowel Dis 2013;19:1322-9. [Crossref] [PubMed]
- Levine A, Rhodes JM, Lindsay JO, et al. Dietary Guidance from the International Organization for the Study of Inflammatory Bowel Disease. Clin Gastroenterol Hepatol 2020;18:1381-92. [Crossref] [PubMed]
- Ashton JJ, Gavin J, Beattie RM. Exclusive enteral nutrition in Crohn's disease: Evidence and practicalities. Clin Nutr 2019;38:80-9. [Crossref] [PubMed]
- Lawley M, Wu JW, Navas-López VM, et al. Global variation in use of enteral nutrition for pediatric Crohn disease. J Pediatr Gastroenterol Nutr 2018;67:e22-9. [Crossref] [PubMed]
- Hart L, Farbod Y, Szamosi JC, et al. Effect of Exclusive Enteral Nutrition and Corticosteroid Induction Therapy on the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Nutrients 2020;12:1691. [Crossref] [PubMed]
- Kaakoush NO, Day AS, Leach ST, et al. Effect of exclusive enteral nutrition on the microbiota of children with newly diagnosed Crohn's disease. Clin Transl Gastroenterol 2015;6:e71 [Crossref] [PubMed]
- Leach S, Mitchell H, Eng W, et al. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn’s disease. Aliment Pharmacol Ther 2008;28:724-33. [Crossref] [PubMed]
- Quince C, Ijaz UZ, Loman N, et al. Extensive Modulation of the Fecal Metagenome in Children With Crohn's Disease During Exclusive Enteral Nutrition. Am J Gastroenterol 2015;110:1718-30. [Crossref] [PubMed]
- Gerasimidis K, Bertz M, Hanske L, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm Bowel Dis 2014;20:861-71. [Crossref] [PubMed]
- Andoh A, Inoue R, Kawada Y, et al. Elemental diet induces alterations of the gut microbial community in mice. J Clin Biochem Nutr 2019;65:118-24. [Crossref] [PubMed]
- Svolos V, Hansen R, Nichols B, et al. Treatment of active Crohn’s disease with an ordinary food-based diet that replicates exclusive enteral nutrition. Gastroenterology 2019;156:1354-67.e6. [Crossref] [PubMed]
- Levine A, Wine E, Assa A, et al. Crohn’s disease exclusion diet plus partial enteral nutrition induces sustained remission in a randomized controlled trial. Gastroenterology 2019;157:440-50.e8. [Crossref] [PubMed]
- Boneh RS, Van Limbergen J, Wine E, et al. Dietary Therapies Induce Rapid Response and Remission in Pediatric Patients With Active Crohn’s Disease. Clin Gastroenterol Hepatol 2021;19:752-9. [Crossref] [PubMed]
- Hillman ET, Lu H, Yao T, et al. Microbial ecology along the gastrointestinal tract. Microbes Environ 2017;32:300-13. [Crossref] [PubMed]
- Tuddenham S, Sears CL. The intestinal microbiome and health. Curr Opin Infect Dis 2015;28:464. [Crossref] [PubMed]
- Valcheva R, Dieleman LA. Prebiotics: Definition and protective mechanisms. Best Pract Res Clin Gastroenterol 2016;30:27-37. [Crossref] [PubMed]
- Williams BA, Grant LJ, Gidley MJ, et al. Gut fermentation of dietary fibres: physico-chemistry of plant cell walls and implications for health. Int J Mol Sci 2017;18:2203. [Crossref] [PubMed]
- Stephen AM, Champ MM-J, Cloran SJ, et al. Dietary fibre in Europe: current state of knowledge on definitions, sources, recommendations, intakes and relationships to health. Nutr Res Rev 2017;30:149-90. [Crossref] [PubMed]
- Hamaker BR, Tuncil YE. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol 2014;426:3838-50. [Crossref] [PubMed]
- Cockburn DW, Koropatkin NM. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol 2016;428:3230-52. [Crossref] [PubMed]
- Patnode ML, Beller ZW, Han ND, et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 2019;179:59-73.e13. [Crossref] [PubMed]
- Chen T, Long W, Zhang C, et al. Fiber-utilizing capacity varies in Prevotella-versus Bacteroides-dominated gut microbiota. Sci Rep 2017;7:2594. [Crossref] [PubMed]
- Jeffery IB, Claesson MJ, O'toole PW, et al. Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol 2012;10:591-2. [Crossref] [PubMed]
- Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207. [Crossref] [PubMed]
- Wu GD, Bushmanc FD, Lewis JD. Diet, the human gut microbiota, and IBD. Anaerobe 2013;24:117-20. [Crossref] [PubMed]
- Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 2008;3:417-27. [Crossref] [PubMed]
- Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016;14:20-32. [Crossref] [PubMed]
- Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol 2018;9:2247. [Crossref] [PubMed]
- Brown AC, Rampertab SD, Mullin GE. Existing dietary guidelines for Crohn’s disease and ulcerative colitis. Expert Rev Gastroenterol Hepatol 2011;5:411-25. [Crossref] [PubMed]
- Owczarek D, Rodacki T, Domagała-Rodacka R, et al. Diet and nutritional factors in inflammatory bowel diseases. World J Gastroenterol 2016;22:895. [Crossref] [PubMed]
- Shinde T, Perera AP, Vemuri R, et al. Synbiotic supplementation containing whole plant sugar cane fibre and probiotic spores potentiates protective synergistic effects in mouse model of IBD. Nutrients 2019;11:818. [Crossref] [PubMed]
- Faghfoori Z, Navai L, Shakerhosseini R, et al. Effects of an oral supplementation of germinated barley foodstuff on serum tumour necrosis factor-α, interleukin-6 and-8 in patients with ulcerative colitis. Ann Clin Biochem 2011;48:233-7. [Crossref] [PubMed]
- Fritsch J, Garces L, Quintero MA, et al. Low-fat, High-fiber Diet Reduces Markers of Inflammation and Dysbiosis and Improves Quality of Life in Patients With Ulcerative Colitis. Clin Gastroenterol Hepatol 2020; Epub ahead of print. [Crossref] [PubMed]
- Kiszonas AM, Fuerst EP, Morris CF. Wheat arabinoxylan structure provides insight into function. Cereal Chem 2013;90:387-95. [Crossref]
- Amodo R, Neukom H. Minor constituents of wheat flour: the pentosan. New Approaches to Research on Cereal Carbohydrates 1985:241-51.
- Dornez E, Gebruers K, Delcour JA, et al. Grain-associated xylanases: occurrence, variability, and implications for cereal processing. Trends Food Sci Technol 2009;20:495-510. [Crossref]
- Mendis M, Leclerc E, Simsek S. Arabinoxylan hydrolyzates as immunomodulators in lipopolysaccharide-induced RAW264. 7 macrophages. Food Funct 2016;7:3039-45. [Crossref] [PubMed]
- Lu ZX, Walker KZ, Muir JG, et al. Arabinoxylan fiber, a byproduct of wheat flour processing, reduces the postprandial glucose response in normoglycemic subjects. Am J Clin Nutr 2000;71:1123-8. [Crossref] [PubMed]
- Henry RJ. A comparison of the non-starch carbohydrates in cereal grains. J Sci Food Agric 1985;36:1243-53. [Crossref]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012;489:242-9. [Crossref] [PubMed]
- Cooper-Bribiesca B, Navarro-Ocaña A, Díaz-Ruiz G, et al. Lactic acid fermentation of arabinoxylan from nejayote by Streptococcus infantarius ssp. infantarius 25124 isolated from pozol. Front Microbiol 2018;9:3061. [Crossref] [PubMed]
- Pastell H, Westermann P, Meyer AS, et al. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed fecal microbiota. J Agric Food Chem 2009;57:8598-606. [Crossref] [PubMed]
- Kanauchi O, Mitsuyama K, Homma T, et al. Treatment of ulcerative colitis patients by long-term administration of germinated barley foodstuff: multi-center open trial. Int J Mol Med 2003;12:701-4. [Crossref] [PubMed]
- Van den Abbeele P, Gérard P, Rabot S, et al. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ Microbiol 2011;13:2667-80. [Crossref] [PubMed]
- Neyrinck AM, Possemiers S, Druart C, et al. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One 2011;6:e20944 [Crossref] [PubMed]
- James SL, Christophersen CT, Bird AR, et al. Abnormal fibre usage in UC in remission. Gut 2015;64:562-70. [Crossref] [PubMed]
- Mendis M, Leclerc E, Simsek S. Arabinoxylan hydrolyzates as immunomodulators in Caco-2 and HT-29 colon cancer cell lines. Food Funct 2017;8:220-31. [Crossref] [PubMed]
- Akramienė D, Kondrotas A, Didžiapetrienė J, et al. Effects of ß-glucans on the immune system. Medicina 2007;43:597. [Crossref] [PubMed]
- Lam KL, Keung HY, Ko KC, et al. In vitro fermentation of beta-glucans and other selected carbohydrates by infant fecal inoculum: An evaluation of their potential as prebiotics in infant formula. Bioactive Carbohydrates and Dietary Fibre 2018;14:20-4. [Crossref]
- Zhao J, Cheung PC. Fermentation of β-glucans derived from different sources by bifidobacteria: Evaluation of their bifidogenic effect. J Agric Food Chem 2011;59:5986-92. [Crossref] [PubMed]
- El Khoury D, Cuda C, Luhovyy B, et al. Beta glucan: health benefits in obesity and metabolic syndrome. J Nutr Metab 2012;2012:851362 [Crossref] [PubMed]
- Vetvicka V, Gover O, Hayby H, et al. Immunomodulating Effects Exerted by Glucans Extracted from the King Oyster Culinary-Medicinal Mushroom Pleurotus eryngii (Agaricomycetes) Grown in Substrates Containing Various Concentrations of Olive Mill Waste. Int J Med Mushrooms 2019;21:765-81. [Crossref] [PubMed]
- Mitmesser S, Combs M. Prebiotics: Inulin and other oligosaccharides. The microbiota in gastrointestinal pathophysiology. Elsevier; 2017. p. 201-8.
- Valcheva R, Koleva P, Martínez I, et al. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes 2019;10:334-57. [Crossref] [PubMed]
- Dewulf EM, Cani PD, Claus SP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013;62:1112-21. [Crossref] [PubMed]
- Dongowski G, Lorenz A, Anger H. Degradation of pectins with different degrees of esterification by Bacteroides thetaiotaomicron isolated from human gut flora. Appl Environ Microbiol 2000;66:1321-7. [Crossref] [PubMed]
- Sahasrabudhe NM, Beukema M, Tian L, et al. Dietary fiber pectin directly blocks Toll-like receptor 2–1 and prevents doxorubicin-induced ileitis. Front Immunol 2018;9:383. [Crossref] [PubMed]
- Tian L, Scholte J, Borewicz K, et al. Effects of pectin supplementation on the fermentation patterns of different structural carbohydrates in rats. Mol Nutr Food Res 2016;60:2256-66. [Crossref] [PubMed]
- Luis AS, Briggs J, Zhang X, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 2018;3:210-9. [Crossref] [PubMed]
- Larsen N, Bussolo de Souza C, Krych L, et al. Potential of pectins to beneficially modulate the gut microbiota depends on their structural properties. Front Microbiol 2019;10:223. [Crossref] [PubMed]
- Kaur SJ, Gupta VK. Production of pectinolytic enzymes pectinase and pectin lyase by Bacillus subtilis SAV-21 in solid state fermentation. Ann Microbiol 2017;67:333-42. [Crossref]
- Kuivanen J, Biz A, Richard P. Microbial hexuronate catabolism in biotechnology. AMB Express 2019;9:16. [Crossref] [PubMed]
- Dittoe DK, Barabote RD, Rothrock MJ, et al. Assessment of a Potential Role of Dickeya dadantii DSM 18020 as a Pectinase Producer for Utilization in Poultry Diets Based on in silico Analyses. Front Microbiol 2020;11:751. [Crossref] [PubMed]
- Benoit I, Coutinho PM, Schols HA, et al. Degradation of different pectins by fungi: correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics 2012;13:321. [Crossref] [PubMed]
- Armstrong H, Bording-Jorgensen M, Wine E. The Multifaceted Roles of Diet, Microbes and Metabolites in Cancer. Cancers 2021;13:767. [Crossref] [PubMed]
- Kim Y, Hwang SW, Kim S, et al. Dietary cellulose prevents gut inflammation by modulating lipid metabolism and gut microbiota. Gut Microbes 2020;11:944-61. [Crossref] [PubMed]
- Mills E, O’Neill LA. Succinate: a metabolic signal in inflammation. Trends in cell biology 2014;24:313-20. [Crossref] [PubMed]
- Davis SR, Cousins RJ. Metallothionein expression in animals: a physiological perspective on function. J Nutr 2000;130:1085-8. [Crossref] [PubMed]

王亮
徐州医科大学。博士,教授。本科毕业于中南大学湘雅医学院,博士毕业于西澳大学医学院病理和医学检验学院,加拿大和澳大利亚博士后,荣获“澳中幽门螺杆菌研究学者”奖,入选江苏省双创博士以及青蓝工程,发表SCI论文50余篇。(更新时间:2021/8/12)
(本译文仅供学术交流,实际内容请以英文原文为准。)
Cite this article as: Dijk S, Armstrong H, Valcheva R, Wine E. Paediatric IBD: the host, diet & microbes in pathogenesis & treatment: a narrative review. Dig Med Res 2021;4:6.


