
Combined olaparib and oxaliplatin inhibits tumor proliferation by cell cycle arrest and cell apoptosis in XRCC2-defecient colorectal cancer
Introduction
Colorectal cancer (CRC) is a common malignant tumor of digestive tract, and its morbidity and mortality maintain an upward trend in the world (1,2). With the rapid development of our social economy, the residents of western-style diet and environmental pollution, CRC incidence will still continuous increase in a period time, become one of the most common malignant tumor in endanger to people's health in our country (3). At present, the targeted therapy of CRC has made significant progress, but still has not achieved satisfactory efficacy, and therefore better targeted drugs are needed.
Chemotherapy is one of the middle-late main treatment for CRC. The cytotoxic drug, oxaliplatin, is a first-line chemotherapy drug for CRC beginning in 2000 (4). It not only improves the objective remission rate but resection rate of Colorectal metastasis tumor (mainly for liver metastasis), and also has extended the overall survival (OS) (5,6). However, tumor intrinsic or acquired drug resistance can still against oxaliplatin killing effect. In the clinical work, we found that the sensitivity of the CRC patients on oxaliplatin chemotherapy varies widely, Some patients are insensitive or even tolerant to chemotherapy (7). Therefore, it is an urgent problem to further study and explore the combination of molecular targeted drugs and chemotherapy to provide safe and effective chemotherapeutic sensitizing drugs for clinical use.
Oxaliplatin is a third-generation platinum analog of the 1,2-diaminocyclohexane family, mainly forms platin-DNA admixture through binding with intracellular DNA, causing DNA damage and resulting in cell death to achieve the purpose of treatment (4,8). However, tumor cells can activate their own DNA damage repair mechanism for damage repair and thus generate chemotherapy resistance. DNA damage repair mainly includes homologous recombination (HR) and nonhomologous end joining (NHEJ) (9-12). Oxaliplatin can induce DNA cross-links and result in DNA double-strand breaks (DSBs) (13,14). After DSBs are induced by irradiation or cytotoxic drugs, H2AX is rapidly phosphorylated. In this case, there is always a considerable number of gamma histone variant H2AX (γ-H2AX) formed per DSB (15).
According to the synthetic lethal theory, both DNA damage repair methods must be inhibited in order to specifically killing tumor cells (16,17). XRCC2 is a key protein involved in HR repair and has been proved in previous studies to be able to predict the efficacy evaluation of CRC radiotherapy (18,19). Poly(ADP-ribose) polymerase 1 (PARP1 has an influence in NHEJ repair and is essential for the outcome of PARP1 inhibitor monotherapy (20). Inhibitors of PARP1 augment the levels of persistent single-strand breaks that lead to DNA DSBs upon replication (21). Our object of this research was to observe the features of the sensitizing impacts of a PARP1 inhibitor on oxaliplatin treatment for XRCC2 deficient CRC for potential clinical application.
Methods
Cell culture and drugs
Human CRC cell line SW480 were mainly derived from the American Type Culture Collection (Manassas, VA, USA). Cells were cultivated at 37 °C with 5% CO2 inRPMI-1640 medium (Gibco, MA, USA), 100 U/mL penicillin as well as 100 µg/mL streptomycin. Olaparib (AZD2281) originated from Selleckchem (Hous-ton, TX, USA) and oxaliplatin stemmed from Hengrui Medicine Co., Ltd. (Jiangsu, China).
Vector and retroviral infection
Several plasmids with lentivirus as the main component were selected. Each plasmid has a short hairpin RNA (ShRNA), which was mainly purchased from RiboBio Co., Ltd. (Guangzhou, China). Their main function is to target human XRCC2. The main function of nonsilencing (vector) shRNAs were to serve as negative control. Human XRCC2 targeted shRNAs infected SW480 cells (22,23). Stable clones will only be obtained after cells transfect SW480 cells in 6-well plate, each plasmid weighed 2 µg. After 48 hours, we will screen them for antibiotics with 2.0 µg/mL of puromycin. After 10 days, multiple clones from the same transfection were mixed together and was grown under the selection of puromycin. Finally, we need to use quantitative RT-PCR and western blots to detect the knockdown efficiency of XRCC2.
Western blotting
Western blotting was carried out based on previously described (24). The antibody sources were as follows: XRCC2 (1:1,000, ab180752), cyclin B1 (1:5,000, ab32053), caspase-3 (1:500, ab13847), caspase-9 (1:1,000, ab202068), BCL-2 (1:500, ab32124), and GADPH (1:2,500, ab9485) were bought from Abcam Inc. Goatanti-rabbit IgG H&L (1:2,000, ab6721, Abcam) was regarded as a second antibody.
Quantitative real-time PCR (qRT-PCR)
As we described earlier, we collected total RNA and carried out qRT-PCR (22). Real-time quantitative PCR was conducted based on SYBR Green I (Invitrogen) having an ABI PRISM 7500 system (Applied Bio systems, Foster City, CA, USA). The housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was employed as an internal control. All experiments were performed at least in triplicate.
Cell proliferation detection
Based on the manufacturer’s guidance, cell proliferation detection was estimated with the help of the Cell Counting Kit-8 (CCK-8) cell proliferation kit (Dojindo Laboratories, Kumamoto, Japan). In a few words, cells were classified into four parts: controls; olaparib (1 µmol/L); oxaliplatin (20 µg/mL); as well as olaparib (1 µmol/L) + oxaliplatin (20 µg/mL). Cells were inoculated into 96-well plates under the condition of 5×103 cells/well and 100 µL of complete medium, and then cultured under normal conditions. We will set a fixed time to incubate the cells with 100 µL of rpmi-1640 medium, plus M+10 µL of CCK 8 reagent, at 37 °C for 2 hours. Then, absorbance was measured on a microplate reader (Bio-Rad, La Jolla, CA, USA), and finally the wavelength was 450 nm. Three independent repeated experiments will be carried out.
Immunofecnecseroul
A previously reported immunofluorescence protocol was used (19,25), with some modifications. After the cells were attached to the chamber slides (Nest, Wuxi, China) on the 24-well plate, they were respectively contacted with olaparib (1 µM) and/or oxaliplatin (10 µM) for 6 hours. At various timepoints following exposure to olaparib and/or oxaliplatin (0, 0.5, 12, 24 and 48 hrs), the cells were washed 3 times with PBS respectively and then fixed in 4% paraformaldehyde at room temperature. Waited for 20 minutes to achieve the effect of cell infiltration. During the whole experiment process, in the first stage, soaked with 0.2% TritonX-100 for 30 min, then rinsed with PBS for 3 times, and finally sealed with 10% goat serum for 60–90 min. In the second stage, coverslips were incubated with anti-γ-H2AX monoclonal antibodies (Cell Signaling Technology). We diluted 1:100 in 1% BSA/PBS/0.1% Tween solution overnight at 4 °C, washed 3 times with PBS, and incubated with Dylight 594 affinity goat anti-mouse IgG (Abbkine, Redlands, CA, USA), the experimental environment was mainly at room temperature in the dark. Waited for 60 minutes, washed with PBS for 3 times, and stained with 2 µg/mL 4',6-diamino-2-phenylindole (DAPI) for 3 min in the dark. Finally, the slide was installed with anti-fading agent and tested with confocal microscope (Zeiss, Germany). In each sample, in a high-power field, the γ-H2AX focus number of each core was counted by an auto mated focus counter, and an average of 100 nuclei were analyzed.
Colony formation assay
Briefly speaking, exponentially growing cells were seeded into 6-wellplates (2,000 cells/well) and cultivated at the condition of 37 °C with 5% CO2 lasting for 10–14 days. Colonies were fixed with 75% ethanol lasting for 30 minutes and then were stained with 0.5% crystal violet (Beyotime, Nanjing, China) for visualization and counting. There were only colonies of more than 50 cells using manual counting. Each group of cells included three wells, and all experiments were repeated three times.
Results
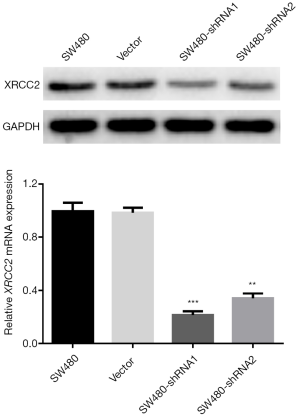
Validation of XRCC2 knockdown in SW480 cells
With the help of lentivirus-mediated shRNAs (XRCC2-sh1 and XRCC2-sh2), expression of XRCC2 was knocked down in SW480 cells (Figure 1A,B). Especially, the SW480 cells that were infected by XRCC2-sh1 exhibited lower expression of XRCC2 weighed against the controls, and these cells were carried out in following experiments, and we named it SW480Sh-XRCC2.
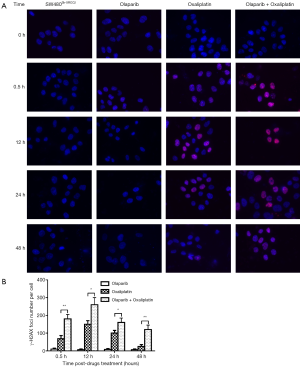
Combination of olaparib and oxaliplatin increased the chemosensitivity of SW480Sh-XRCC2 cell by induces γ-H2AX foci formation
To examine the DSB repair efficiency of combination of olaparib and oxaliplatin in SW480Sh-XRCC2 cells compared to olaparib or oxaliplatin alone, phosphorylation of H2AX (γ-H2AX) was determined. As mentioned earlier, the γ-H2AX lesion did not disappear but persisted after drugs treatment, which reflects that the ability of cells to repair DNA DSBs has been damaged (19). We then analyzed the γ-H2AX lesion by immunofluorescence staining, and then treated it with Olaparib and/or oxaliplatin. We compared the characteristics of SW480Sh-XRCC2 cells treated by olaparib and control cells, and the results showed that there was no difference between them. Therefore, this indicates that olaparib alone did not induce DSBs. Experiments prove that SW480Sh-XRCC2 cells treated with oxaliplatin and olaparib show more γ-H2AX foci than cells treated with oxaliplatin or olaparib alone. However, the experimental results also showed that the γ-H2AX lesion disappeared faster on the cells treated with oxaliplatin were combined with SW480Sh-XRCC2 cells exposed to oxaliplatin at 24 hours and 48 hours after drug treatment to use olaparib (Figure 2; P<0.01). These results show that olaparib may increase the sensitivity of SW480Sh-XRCC2 cells to oxaliplatin by enhancing induced DNA DSBs.
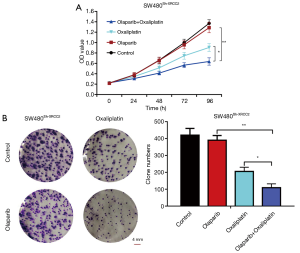
Combination of olaparib and oxaliplatin inhibits SW480Sh-XRCC2 cell proliferation
At 24, 48, 72 and 96 hours after administration, we assessed the proliferation rate of olaparib treated cells. The effect of olaparib combined with oxaliplatin on cell proliferation is dramatically lower than that of cells treated with oxaliplatin alone or olaparib alone (Figure 3A; P<0.05 and P<0.01). Moreover, a clonogenic assay indicated that, compared with oxaliplatin group or olaparib group, olaparib combined with oxaliplatin group showed obviously declined colony formation capacity after 10–14 of days conventional culture (Figure 3B; P<0.05 and P<0.01). To sum up, olaparib can improve the sensitivity of SW480Sh-XRCC2 cells to oxaliplatin.
Combination of olaparib and oxaliplatin induces G2/M arrest in SW480Sh-XRCC2 cell
The flow cytometry assay gave the description that there was no obvious difference in the proportion of SW480Sh-XRCC2 cells in G2/M-phase between the cells treated with olaparib and the control cells showing that olaparib alone did not induce G2/M arrest. Additionally, SW480Sh-XRCC2 cells treated with the combination of olaparib and oxaliplatin have more G2/M phase cells than cells treated by oxaliplatin alone or olaparib alone (Figure 4A; P<0.05 and P<0.01). In addition, biochemical markers of cell cycle were detected. Western blot analysis showed that the protein level of G2-phase related cyclinB1 was up-regulated (Figure 4B). Collectively, these results suggest that olaparib helps oxaliplatin to arrest SW480Sh-XRCC2 cells in G2/M-phase.
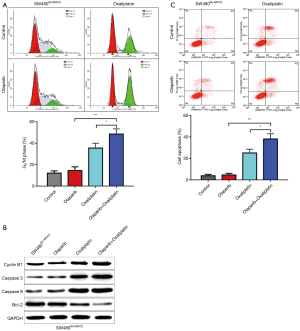
Combination of olaparib and oxaliplatin enhanced apoptosis in SW480Sh-XRCC2 cell
To quantify the apoptotic effects of combination of olaparib and oxaliplatin in SW480Sh-XRCC2 cell, cell apoptosis was assessed with the help of flow cytometry. As the description in Figure 4C, the combination of olaparib and oxaliplatin treated in SW480Sh-XRCC2 cell exhibited a significant increase in apoptosis compared with the olaparib alone or oxaliplatin alone treated (P<0.05 and P<0.01). In an effort to examine the possible mechanisms of this observed augment in drug sensitivity, biochemical markers of apoptosis were tested. These markers were as follows: cleaved caspase-9, cleaved caspase-3, as well as anti-apoptosis (Bcl-2). Higher levels of cleaved caspase-9 and cleaved caspase-3 were tested in combination of olaparib and oxaliplatin treated SW480Sh-XRCC2 cells compared to the olaparib alone or oxaliplatin alone treated cells was conducted to the cells (Figure 4B). In contrary to the previous hypothesis, an obvious decline in the levels of Bcl-2 were examined in the combination of olaparib and oxaliplatin treated SW480Sh-XRCC2 cells (Figure 4B). Taken together, these findings indicate that olaparib may enhanced the chemosensitivity of oxaliplatin by induced apoptosis.
Discussion
Previous researches show that increasing XRCC2 expression advance the invasive behaviors along with metastasis processes of CRC (22). Our previous studies have found that patients with high XRCC2 expression are sensitive to radiotherapy and chemotherapy (19,22). However, Can drugs increase the sensitivity of XRCC2-deficient CRC patients to chemotherapy and radiotherapy? A variety of PARP1 inhibitors have been investigated in recent years. Olaparib can be defined as a bioavailable inhibitor of PARP1, and some clinical trial based on olaparib has been recorded. Studies have shown that after oral administration of 100 or 200 mg, the plasma concentration of olaparib can reach 11.5 mL, but the two dosages will not cause dose-limiting toxicity (26). When investigating the impact of a PARP1 inhibitor, Vilar et al. (27) discover that the MRE11 cell line combined with harboring biallelic mutations were more sensitive to veliparib than wild-type cell lines. In the whole process of this study, we persist in using 1.0 µM olaparib (under the guidance of the results of our previous survey). We mainly compared the influence of olaparib and/or oxaliplatin on XRCC2 gene knockdown SW480 cells. After comparison, we discovered that olaparib has cytotoxic impact contrasted to the control group. Besides, the research results also show that the combination of oxaliplatin and olaparib can significantly improve the killing effect of oxaliplatin. Thus, for the treatment of patients with XRCC2 deficiency, the combination of PARP inhibitor and oxaliplatin has broad clinical application prospects.
Researchers have the same view that γ-H2AX lesion is a good sign of DSBs. In this case, we can use anti-γ-H2AX antibody to count the lesions in cells, so as to achieve the purpose of detecting DSBs. Therefore, we detected γ-H2AX levels after treatment with the combination of olaparib and oxaliplatin (28-30). Although Chan et al. (31) discovered that PARP inhibition alone did not induce a statistically obvious increase in γ-H2AX expression in vitro or in vivo. Immunofluorescence results indicts that olaparib connected with oxaliplatin generated the most foci among all the groups, and the speed of repaired of DSBs in olaparib combined with oxaliplatin group was slowest compared with oxaliplatin group. These results shown that the sensitizing impact of olaparib for oxaliplatin may be connected with the induction of DSBs.
In the present research, we discovered that combination of olaparib and oxaliplatin may reduce the XRCC2-deficient CRC cell proliferation rate as well as colony formation capacity rather than oxaliplatin or olaparib alone (P<0.05 and P<0.01), and suggested that olaparib as well as oxaliplatin have a synergistic impact on inhibit the proliferation function in XRCC2-deficient CRC. Arango et al. (32) discovered that exposing of colon cancer cells to oxaliplatin leaded to G2/M arrest and apoptosis. Our outcomes shown that oxaliplatin alone can contribute to G2/M arrest. Moreover, cells with the integration of olaparib and oxaliplatin have more G2/M-phase cells than oxaliplatin or olaparib alone (P<0.05 and P<0.01). Our Western blot outcomes predict that cyclin B1 protein level were increased obviously in combination of olaparib and oxaliplatin group. These outcomes predict that olaparib and oxaliplatin have a synergistic impact on G2/M arrest. In addition, olaparib plays an important role to oxaliplatin in arresting XRCC2-deficient CRC cells in G2/M-phase. Flow cytometry suggested that combination of olaparib and oxaliplatin increased XRCC2-deficient CRC cell apoptosis rather than oxaliplatin or olaparib alone (P<0.05 and P<0.01), and confirmed the protein levels. These outcomes show that combination of olaparib and oxaliplatin may influence cell proliferation of XRCC2-deficient CRC cells with the aid of cell cycle arrest along with cell apoptosis.
In conclusion, the results of this study demonstrate that olaparib and oxaliplatin have a synergistic effect on the capacity to repair DNA DSBs and proliferation ability of XRCC2-deficient CRC cell, which caused more γ-H2AX formation, more G2/M phase cells arrest and more cells apoptosis. Our findings suggest that combined treatment with olaparib and oxaliplatin could be a desirable method for clinical chemotherapy in XRCC2-defecient CRC, although further investigation is needed.
Acknowledgments
Funding: This work was supported by the Science and Technology Planning Project of Guangdong Province, China (2017A020215014); Medical Scientific Research Foundation of Guangdong Province, China (A2017274).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/dmr.2019.12.02). YH serves as the Editor-in-Chief of Digestive Medicine Research. CZ serves as an unpaid Associate Editor-in-Chief of Digestive Medicine Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-93. [Crossref] [PubMed]
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Mathé G, Kidani Y, Segiguchi M, et al. Oxalato-platinum or 1-OHP, a third-generation platinum complex: an experimental and clinical appraisal and preliminary comparison with cis-platinum and carboplatinum. Biomed Pharmacother 1989;43:237-50. [Crossref] [PubMed]
- Rothenberg ML. Efficacy of oxaliplatin in the treatment of colorectal cancer. Oncology (Williston Park) 2000;14:9-14. [PubMed]
- Culy CR, Clemett D, Wiseman LR, et al. A review of its pharmacological properties and clinical efficacy in metastatic colorectal cancer and its potential in other malignancies. Drugs 2000;60:895-924. [Crossref] [PubMed]
- Matsuo H, Nakanishi M, Arita T, et al. Tolerability of Oxaliplatin-Based Adjuvant Chemotherapy for Patients with Colorectal Cancer. Gan To Kagaku Ryoho 2015;42:2112-4. [PubMed]
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 2014;740:364-78. [Crossref] [PubMed]
- Adachi N, Iiizumi S, So S, et al. Genetic evidence for involvement of two distinct nonhomologous end-joining pathways in repair of topoisomerase II-mediated DNA damage. Biochem Biophys Res Commun 2004;318:856-61. [Crossref] [PubMed]
- Srivastava M, Nambiar M, Sharma S, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 2012;151:1474-87. [Crossref] [PubMed]
- Dilley RL, Verma P, Cho NW, et al. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016;539:54-58. [Crossref] [PubMed]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 2008;18:99-113. [Crossref] [PubMed]
- Chiu SJ, Lee YJ, Hsu TS, et al. Oxaliplatin-induced gamma-H2AX activation via both p53-dependent and -independent pathways but is not associated with cell cycle arrest in human colorectal cancer cells. Chem Biol Interact 2009;182:173-82. [Crossref] [PubMed]
- Woynarowski JM, Faivre S, Herzig MC, et al. Oxaliplatin-induced damage of cellular DNA. Mol Pharmacol 2000;58:920-7. [Crossref] [PubMed]
- Rogakou EP, Nieves-Neira W, Boon C, et al. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem 2000;275:9390-5. [Crossref] [PubMed]
- O'Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet 2017;18:613-23. [Crossref] [PubMed]
- Lok BH, Carley AC, Tchang B, et al. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 2013;32:3552-8. [Crossref] [PubMed]
- Gil Del Alcazar CR, Todorova PK, Habib AA, et al. Augmented HR Repair Mediates Acquired Temozolomide Resistance in Glioblastoma. Mol Cancer Res 2016;14:928-40. [Crossref] [PubMed]
- Qin CJ, Song XM, Chen ZH, et al. XRCC2 as a predictive biomarker for radioresistance in locally advanced rectal cancer patients undergoing preoperative radiotherapy. Oncotarget 2015;6:32193-204. [Crossref] [PubMed]
- Tentori L, Muzi A, Dorio AS, et al. MSH3 expression does not influence the sensitivity of colon cancer HCT116 cell line to oxaliplatin and poly(ADP-ribose) polymerase (PARP) inhibitor as monotherapy or in combination. Cancer Chemother Pharmacol 2013;72:117-25. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Xu K, Song X, Chen Z, et al. XRCC2 promotes colorectal cancer cell growth, regulates cell cycle progression, and apoptosis. Medicine (Baltimore) 2014;93:e294. [Crossref] [PubMed]
- Vilar E, Bartnik CM, Stenzel SL, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res 2011;71:2632-42. [Crossref] [PubMed]
- Ren H, Chen Z, Yang L, et al. Apolipoprotein C1 (APOC1) promotes tumor progression via MAPK signaling pathways in colorectal cancer. Cancer Manag Res 2019;11:4917-30. [Crossref] [PubMed]
- Zheng Z, Ng WL, Zhang X, et al. RNAi-mediated targeting of noncoding and coding sequences in DNA repair gene messages efficiently radiosensitizes human tumor cells. Cancer Res 2012;72:1221-8. [Crossref] [PubMed]
- Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123-34. [Crossref] [PubMed]
- Vilar E, Bartnik CM, Stenzel SL, et al. MRE11 deficiency increases sensitivity to poly (ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res 2011;71:2632-42. [Crossref] [PubMed]
- Yoshikawa T, Kashino G, Ono K, et al. Phosphorylated H2AX foci in tumor cells have no correlation with their radiation sensitivities. J Radiat Res 2009;50:151-60. [Crossref] [PubMed]
- Kinner A, Wu W, Staudt C, et al. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008;36:5678-94. [Crossref] [PubMed]
- Kato TA, Nagasawa H, Weil MM, et al. gamma-H2AX foci after low-dose-rate irradiation reveal atm haploinsufficiency in mice. Radiat Res 2006;166:47-54. [Crossref] [PubMed]
- Chan N, Pires IM, Bencokova Z, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res 2010;70:8045-54. [Crossref] [PubMed]
- Arango D, Wilson AJ, Shi Q, et al. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br J Cancer 2004;91:1931-46. [Crossref] [PubMed]
Cite this article as: Ren H, Wu W, Li M, Yang S, Chen J, Chen H, Li L, Zhang C, He Y. Combined olaparib and oxaliplatin inhibits tumor proliferation by cell cycle arrest and cell apoptosis in XRCC2-defecient colorectal cancer. Dig Med Res 2019;2:41.


